Evidence of active methanogen communities in shallow sediments of the sonora margin cold seeps
- PMID: 25769831
- PMCID: PMC4407212
- DOI: 10.1128/AEM.00147-15
Evidence of active methanogen communities in shallow sediments of the sonora margin cold seeps
Abstract
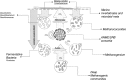
In the Sonora Margin cold seep ecosystems (Gulf of California), sediments underlying microbial mats harbor high biogenic methane concentrations, fueling various microbial communities, such as abundant lineages of anaerobic methanotrophs (ANME). However, the biodiversity, distribution, and metabolism of the microorganisms producing this methane remain poorly understood. In this study, measurements of methanogenesis using radiolabeled dimethylamine, bicarbonate, and acetate showed that biogenic methane production in these sediments was mainly dominated by methylotrophic methanogenesis, while the proportion of autotrophic methanogenesis increased with depth. Congruently, methane production and methanogenic Archaea were detected in culture enrichments amended with trimethylamine and bicarbonate. Analyses of denaturing gradient gel electrophoresis (DGGE) fingerprinting and reverse-transcribed PCR-amplified 16S rRNA sequences retrieved from these enrichments revealed the presence of active methylotrophic Methanococcoides burtonii relatives and several new autotrophic Methanogenium lineages, confirming the cooccurrence of Methanosarcinales and Methanomicrobiales methanogens with abundant ANME populations in the sediments of the Sonora Margin cold seeps.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Boetius A, Wenzhofer F. 2013. Seafloor oxygen consumption fuelled by methane from cold seeps. Nat Geosci 6:725–734. doi:10.1038/ngeo1926. - DOI
-
- Hinrichs K-U, Boetius A. 2002. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry, p 457–477. In Wefer G, Billett D, Hebbeln D, Jørgensen BB, Schlüter M, Van Weering T (ed), Ocean margin systems. Springer-Verlag, Berlin, Germany.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

