Multiple HIV-1 infections with evidence of recombination in heterosexual partnerships in a low risk Rural Clinical Cohort in Uganda
- PMID: 21239033
- PMCID: PMC3041926
- DOI: 10.1016/j.virol.2010.12.025
Multiple HIV-1 infections with evidence of recombination in heterosexual partnerships in a low risk Rural Clinical Cohort in Uganda
Abstract
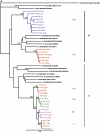
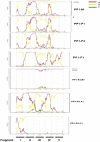
We report on the frequency of multiple infections, generation of recombinants and consequences on disease progression in 35 HIV-1 infected individuals from 7 monogamous and 6 polygamous partnerships within a Rural Clinical Cohort in Uganda. The env-C2V3, gag-p24 and pol-IN genes were sequenced. Single genome amplified half genome sequences were used to map recombination breakpoints. Three participants were dually infected with subtypes A and D, one case with subtype A and A/D recombinant and the fifth with 2 phylogenetically distinct A/D recombinants. Occurrence of A/D recombination was observed in two multiple infected individuals. Rate of late stage WHO events using Cox regression was 3 times greater amongst multiple infected compared to singly infected individuals (hazard ratio 3.35; 95% CI 1.09, 10.3; p=0.049). We have shown that polygamous relationships involving subtype discordant partnerships was a major contributor of multiple infections with generation of inter subtype recombinants in our cohort.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures









References
-
- Altfeld M., Allen T.M., Yu X.G., Johnston M.N., Agrawal D., Korber B.T., Montefiori D.C., O'Connor D.H., Davis B.T., Lee P.K., Maier E.L., Harlow J., Goulder P.J., Brander C., Rosenberg E.S., Walker B.D. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature. 2002;420(6914):434–439. - PubMed
-
- Bezemer D., van Sighem A., de Wolf F., Cornelissen M., van der Kuyl A.C., Jurriaans S., van der Hoek L., Prins M., Coutinho R.A., Lukashov V.V. Combination antiretroviral therapy failure and HIV super-infection. AIDS. 2008;22(2):309–311. - PubMed
-
- Casado C., Pernas M., Alvaro T., Sandonis V., Garcia S., Rodriguez C., del Romero J., Grau E., Ruiz L., Lopez-Galindez C. Coinfection and superinfection in patients with long-term, nonprogressive HIV-1 disease. J. Infect. Dis. 2007;196(6):895–899. - PubMed
-
- Delwart E.L., Busch M.P., Kalish M.L., Mosley J.W., Mullins J.I. Rapid molecular epidemiology of human immunodeficiency virus transmission. AIDS Res. Hum. Retroviruses. 1995;11(9):1081–1093. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

