A glimpse of the prokaryotic diversity of the Large Aral Sea reveals novel extremophilic bacterial and archaeal groups
- PMID: 31058468
- PMCID: PMC6741134
- DOI: 10.1002/mbo3.850
A glimpse of the prokaryotic diversity of the Large Aral Sea reveals novel extremophilic bacterial and archaeal groups
Abstract
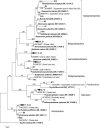
During the last five decades, the Aral Sea has gradually changed from a saline water body to a hypersaline lake. Microbial community inhabiting the Aral Sea has been through a succession and continuous adaptation during the last 50 years of increasing salinization, but so far, the microbial diversity has not been explored. Prokaryotic diversity of the Large Aral Sea using cultivation-independent methods based on determination of environmental 16S rRNA gene sequences revealed a microbial community related to typical marine or (hyper) saline-adapted Bacteria and Archaea. The archaeal sequences were phylogenetically affiliated with the order Halobacteriales, with a large number of operational taxonomic units constituting a novel cluster in the Haloferacaceae family. Bacterial community analysis indicated a higher diversity with representatives belonging to Proteobacteria, Actinobacteria and Bacteroidetes. Many members of Alphaproteobacteria and Gammaproteobacteria were affiliated with genera like Roseovarius, Idiomarina and Spiribacter which have previously been found in marine or hypersaline waters. The majority of the phylotypes was most closely related to uncultivated organisms and shared less than 97% identity with their closest match in GenBank, indicating a unique community structure in the Large Aral Sea with mostly novel species or genera.
Keywords: archaea; bacteria; halophiles; phylogeny; prokaryotic diversity; salt lake.
© 2019 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures


References
-
- Aripov, T. F. , Kukanova, S. I. , Zaynitdinova, L. I. , & Tashpulatov, J. J. (2016). Microorganisms of the extreme zones of the Southern Aral Sea Region. BioTechnology: An Indian Journal, 12(5), 1–7.
-
- Baas‐Becking, L. G. M. (1934). Geobiologie of inleiding tot de milieukunde. The Hague, The Nederlands: Van Stockum and Zoon.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

