Functional identification of the Proteus mirabilis core lipopolysaccharide biosynthesis genes
- PMID: 20622068
- PMCID: PMC2937370
- DOI: 10.1128/JB.00494-10
Functional identification of the Proteus mirabilis core lipopolysaccharide biosynthesis genes
Abstract
In this study, we report the identification of genes required for the biosynthesis of the core lipopolysaccharides (LPSs) of two strains of Proteus mirabilis. Since P. mirabilis and Klebsiella pneumoniae share a core LPS carbohydrate backbone extending up to the second outer-core residue, the functions of the common P. mirabilis genes was elucidated by genetic complementation studies using well-defined mutants of K. pneumoniae. The functions of strain-specific outer-core genes were identified by using as surrogate acceptors LPSs from two well-defined K. pneumoniae core LPS mutants. This approach allowed the identification of two new heptosyltransferases (WamA and WamC), a galactosyltransferase (WamB), and an N-acetylglucosaminyltransferase (WamD). In both strains, most of these genes were found in the so-called waa gene cluster, although one common core biosynthetic gene (wabO) was found outside this cluster.
Figures


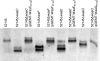
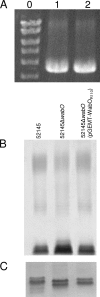
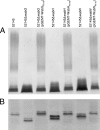


References
-
- Belas, R. 1996. Proteus mirabilis swarmer cell differentiation and urinary tract infection, p. 271-298. In H. L. T. Mobley and J. W. Warren (ed.), Urinary tract infections: molecular pathogenesis and clinical management. ASM Press, Washington, DC.
-
- Belunis, C. J., T. Clementz, S. M. Carty, and C. H. R. Raetz. 1995. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene of Escherichia coli. J. Biol. Chem. 270:27646-27652. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

