Comparative analyses of the bacterial microbiota of the human nostril and oropharynx
- PMID: 20802827
- PMCID: PMC2925076
- DOI: 10.1128/mBio.00129-10
Comparative analyses of the bacterial microbiota of the human nostril and oropharynx
Abstract
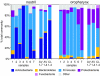
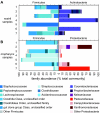
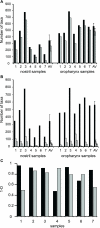
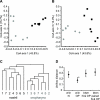
The nose and throat are important sites of pathogen colonization, yet the microbiota of both is relatively unexplored by culture-independent approaches. We examined the bacterial microbiota of the nostril and posterior wall of the oropharynx from seven healthy adults using two culture-independent methods, a 16S rRNA gene microarray (PhyloChip) and 16S rRNA gene clone libraries. While the bacterial microbiota of the oropharynx was richer than that of the nostril, the oropharyngeal microbiota varied less among participants than did nostril microbiota. A few phyla accounted for the majority of the bacteria detected at each site: Firmicutes and Actinobacteria in the nostril and Firmicutes, Proteobacteria, and Bacteroidetes in the oropharynx. Compared to culture-independent surveys of microbiota from other body sites, the microbiota of the nostril and oropharynx show distinct phylum-level distribution patterns, supporting niche-specific colonization at discrete anatomical sites. In the nostril, the distribution of Actinobacteria and Firmicutes was reminiscent of that of skin, though Proteobacteria were much less prevalent. The distribution of Firmicutes, Proteobacteria, and Bacteroidetes in the oropharynx was most similar to that in saliva, with more Proteobacteria than in the distal esophagus or mouth. While Firmicutes were prevalent at both sites, distinct families within this phylum dominated numerically in each. At both sites there was an inverse correlation between the prevalences of Firmicutes and another phylum: in the oropharynx, Firmicutes and Proteobacteria, and in the nostril, Firmicutes and Actinobacteria. In the nostril, this inverse correlation existed between the Firmicutes family Staphylococcaceae and Actinobacteria families, suggesting potential antagonism between these groups.
Figures

References
-
- Wilson M. 2005. Microbial inhabitants of human: their ecology and role in health and disease. Cambridge University Press, Cambridge, United Kingdom
-
- Mertz D., Frei R., Jaussi B., Tietz A., Stebler C., Fluckiger U., Widmer A. F. 2007. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus. Clin. Infect. Dis. 45:475–477 - PubMed
-
- Wertheim H. F., Melles D. C., Vos M. C., van Leeuwen W., Belkum A., Verbrugh H. A., Nouwen J. L. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751–762 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
