Genetic and neutralization sensitivity of diverse HIV-1 env clones from chronically infected patients in China
- PMID: 21325278
- PMCID: PMC3077651
- DOI: 10.1074/jbc.M111.224527
Genetic and neutralization sensitivity of diverse HIV-1 env clones from chronically infected patients in China
Abstract
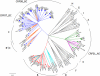
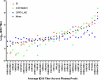
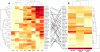
As HIV-1 continues to spread in China from traditional high risk populations to the general public, its genetic makeup has become increasingly complex. However, the impact of these genetic changes on the biological and neutralization sensitivity of the virus is unknown. The current study aims to characterize the genetic, biological, and neutralization sensitivity of HIV-1 identified in China between 2004 and 2007. Based on a total of 107 full-length envelope genes obtained directly from the infected patients, we found that those viruses fell into three major genetic groups: CRF01_AE, subtype B', and subtype C/CRF07_BC/CRF08_BC/B'C. Pseudotyped viruses built upon the viable env genes have demonstrated their substantial variability in mediating viral entry and in sensitivity to neutralization by subtype-specific plasma pools and broadly neutralizing monoclonal antibodies (bnmAb). Many viruses are resistant to one or more bnmAb, including those known to have high potency against diverse viruses from outside China. Sequence and structural analysis has revealed several mechanisms by which these resistant viruses escape recognition from bnmAb. We believe that these results will help us to better understand the impact of genetic diversity on the neutralizing sensitivity of the viruses and to facilitate the design of immunogens capable of eliciting antibodies with potency and breadth similar to those of bnmAb.
Figures

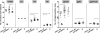
References
-
- Lu L., Jia M., Ma Y., Yang L., Chen Z., Ho D. D., Jiang Y., Zhang L. (2008) Nature 455, 609–611 - PubMed
-
- Ministry of Health of the People's Republic of China (2010) China 2010 UNGASS Country Progress Report (2008–2009), Ministry of Health, Beijing, China
-
- Han X., Dai D., Zhao B., Liu J., Ding H., Zhang M., Hu Q., Lu C., Goldin M., Takebe Y., Zhang L., Shang H. (2010) J. Acquir. Immune Defic. Syndr. 53, Suppl. 1, S27–S33 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

