A customized gene expression microarray reveals that the brittle stem phenotype fs2 of barley is attributable to a retroelement in the HvCesA4 cellulose synthase gene
- PMID: 20530215
- PMCID: PMC2923883
- DOI: 10.1104/pp.110.158329
A customized gene expression microarray reveals that the brittle stem phenotype fs2 of barley is attributable to a retroelement in the HvCesA4 cellulose synthase gene
Abstract
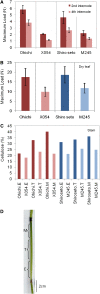
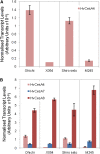
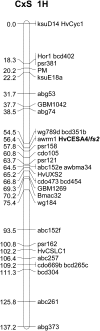
The barley (Hordeum vulgare) brittle stem mutants, fs2, designated X054 and M245, have reduced levels of crystalline cellulose compared with their parental lines Ohichi and Shiroseto. A custom-designed microarray, based on long oligonucleotide technology and including genes involved in cell wall metabolism, revealed that transcript levels of very few genes were altered in the elongation zone of stem internodes, but these included a marked decrease in mRNA for the HvCesA4 cellulose synthase gene of both mutants. In contrast, the abundance of several hundred transcripts changed in the upper, maturation zones of stem internodes, which presumably reflected pleiotropic responses to a weakened cell wall that resulted from the primary genetic lesion. Sequencing of the HvCesA4 genes revealed the presence of a 964-bp solo long terminal repeat of a Copia-like retroelement in the first intron of the HvCesA4 genes of both mutant lines. The retroelement appears to interfere with transcription of the HvCesA4 gene or with processing of the mRNA, and this is likely to account for the lower crystalline cellulose content and lower stem strength of the mutants. The HvCesA4 gene maps to a position on chromosome 1H of barley that coincides with the previously reported position of fs2.
Figures

References
-
- Appenzeller L, Doblin M, Barreiro R, Wang H, Niu X, Kollipara K, Carrigan L, Tomes D, Chapman M, Dhugga KS. (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11: 287–299
-
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 - PubMed
-
- Ballman KV, Grill DE, Oberg AL, Therneau TM. (2004) Faster cyclic Loess: normalizing RNA arrays via linear models. Bioinformatics 20: 2778–2786 - PubMed
-
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300
-
- Bernhardt C, Tierney ML. (2006) Proline-rich cell-wall proteins: building blocks for an expanding cell wall? Hayashi T, , The Science and Lore of the Plant Cell Wall. Brown Walker Press, Boca Raton, FL, pp 164–170
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

