Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis
- PMID: 20709840
- PMCID: PMC2950454
- DOI: 10.1128/AEM.01268-10
Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis
Abstract
The genotyping of numerous isolates of Cryptosporidium parasites has led to the definition of new species and a better understanding of the epidemiology of cryptosporidiosis. A single-locus genotyping method based on the partial sequence of a polymorphic sporozoite surface glycoprotein gene (GP60) has been favored by many for surveying Cryptosporidium parvum and C. hominis populations. Since genetically distinct Cryptosporidium parasites recombine in nature, it is unclear whether single-locus classifications can adequately represent intraspecies diversity. To address this question, we investigated whether multilocus genotypes of C. parvum and C. hominis cluster according to the GP60 genotype. C. hominis multilocus genotypes did not segregate according to this marker, indicating that for this species the GP60 sequence is not a valid surrogate for multilocus typing methods. In contrast, in C. parvum the previously described "anthroponotic" genotype was confirmed as a genetically distinct subspecies cluster characterized by a diagnostic GP60 allele. However, as in C. hominis, several C. parvum GP60 alleles did not correlate with distinct subpopulations. Given the rarity of some C. parvum GP60 alleles in our sample, the existence of additional C. parvum subgroups with unique GP60 alleles cannot be ruled out. We conclude that with the exception of genotypically distinct C. parvum subgroups, multilocus genotyping methods are needed to characterize C. parvum and C. hominis populations. Unless parasite virulence is controlled at the GP60 locus, attempts to find associations within species or subspecies between GP60 and phenotype are unlikely to be successful.
Figures
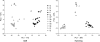
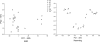
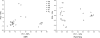

References
-
- Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. - PubMed
-
- Cacciò, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120(Pt. 3):237-244. - PubMed
-
- Cama, V. A., J. M. Ross, S. Crawford, V. Kawai, R. Chavez-Valdez, D. Vargas, A. Vivar, E. Ticona, M. Navincopa, J. Williamson, Y. Ortega, R. H. Gilman, C. Bern, and L. Xiao. 2007. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 196:684-691. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

