Phylogenetic analyses of the polyprotein coding sequences of serotype O foot-and-mouth disease viruses in East Africa: evidence for interserotypic recombination
- PMID: 20731826
- PMCID: PMC2939557
- DOI: 10.1186/1743-422X-7-199
Phylogenetic analyses of the polyprotein coding sequences of serotype O foot-and-mouth disease viruses in East Africa: evidence for interserotypic recombination
Abstract
Background: Foot-and-mouth disease (FMD) is endemic in East Africa with the majority of the reported outbreaks attributed to serotype O virus. In this study, phylogenetic analyses of the polyprotein coding region of serotype O FMD viruses from Kenya and Uganda has been undertaken to infer evolutionary relationships and processes responsible for the generation and maintenance of diversity within this serotype. FMD virus RNA was obtained from six samples following virus isolation in cell culture and in one case by direct extraction from an oropharyngeal sample. Following RT-PCR, the single long open reading frame, encoding the polyprotein, was sequenced.
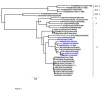
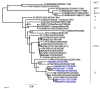
Results: Phylogenetic comparisons of the VP1 coding region showed that the recent East African viruses belong to one lineage within the EA-2 topotype while an older Kenyan strain, K/52/1992 is a representative of the topotype EA-1. Evolutionary relationships between the coding regions for the leader protease (L), the capsid region and almost the entire coding region are monophyletic except for the K/52/1992 which is distinct. Furthermore, phylogenetic relationships for the P2 and P3 regions suggest that the K/52/1992 is a probable recombinant between serotypes A and O. A bootscan analysis of K/52/1992 with East African FMD serotype A viruses (A21/KEN/1964 and A23/KEN/1965) and serotype O viral isolate (K/117/1999) revealed that the P2 region is probably derived from a serotype A strain while the P3 region appears to be a mosaic derived from both serotypes A and O.
Conclusions: Sequences of the VP1 coding region from recent serotype O FMDVs from Kenya and Uganda are all representatives of a specific East African lineage (topotype EA-2), a probable indication that hardly any FMD introductions of this serotype have occurred from outside the region in the recent past. Furthermore, evidence for interserotypic recombination, within the non-structural protein coding regions, between FMDVs of serotypes A and O has been obtained. In addition to characterization using the VP1 coding region, analyses involving the non-structural protein coding regions should be performed in order to identify evolutionary processes shaping FMD viral populations.
Figures


References
-
- Belsham GJ. Translation and Replication of FMDV RNA. In 'Foot and mouth disease virus'. Curr Top Microbiol Immunol. 2005;288:43–70. full_text. - PubMed
-
- Coetzer JAW, Thomsen GR, Tustin RC, Kriek NPJ. Infectious Diseases of Livestock with Special Reference to Southern Africa. Cape Town: Oxford University Press; 1994. Foot-and-mouth disease.
-
- Domingo E, Escarmısa C, Martinez MA, Martinez-Salas E, Mateu MG. Foot-and-mouth disease virus populations are quasispecies. Curr Top Microbiol Immunol. 1992;176:33–47. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

