Prokaryotic diversity in Aran-Bidgol salt lake, the largest hypersaline playa in Iran
- PMID: 22185719
- PMCID: PMC4036037
- DOI: 10.1264/jsme2.me11267
Prokaryotic diversity in Aran-Bidgol salt lake, the largest hypersaline playa in Iran
Abstract
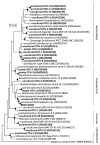
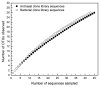
Prokaryotic diversity in Aran-Bidgol salt lake, a thalasohaline lake in Iran, was studied by fluorescence in situ hybridization (FISH), cultivation techniques, denaturing gradient gel electrophoresis (DGGE) of PCR-amplified fragments of 16S rRNA genes and 16S rRNA gene clone library analysis. Viable counts obtained (2.5-4 × 10(6) cells mL(-1)) were similar to total cell abundance in the lake determined by DAPI direct count (3-4×10(7) cells mL(-1)). The proportion of Bacteria to Archaea in the community detectable by FISH was unexpectedly high and ranged between 1:3 and 1:2. We analyzed 101 archaeal isolates and found that most belonged to the genera Halorubrum (55%) and Haloarcula (18%). Eleven bacterial isolates obtained in pure culture were affiliated with the genera Salinibacter (18.7%), Salicola (18.7%) and Rhodovibrio (35.3%). Analysis of inserts of 100 clones from the eight 16S rRNA clone libraries constructed revealed 37 OTUs. The majority (63%) of these sequences were not related to any previously identified taxa. Within this sampling effort we most frequently retrieved phylotypes related to Halorhabdus (16% of archaeal sequences obtained) and Salinibacter (36% of bacterial sequences obtained). Other prokaryotic groups that were abundant included representatives of Haloquadratum, the anaerobic genera Halanaerobium and Halocella, purple sulfur bacteria of the genus Halorhodospira and Cyanobacteria.
Figures




References
-
- Antón J, Llobet-Brossa E, Rodríguez-Valera F, Amann R. Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol. 1999;1:517–523. - PubMed
-
- Benlloch S, Acinas SG, Martínez-Mucia AJ, Rodríguez-Valera F. Description of prokaryotic biodiversity along the salinity gradient of a multipond solar saltern by direct PCR amplification of 16S rDNA. Hydrobiologia. 1996;329:19–31.
-
- Benlloch S, Acinas SG, Antón J, López-López A, Luz SP, Rodríguez-Valera F. Archaeal biodiversity in crystallizer ponds from a solar saltern: culture versus PCR. Microb Ecol. 2001;41:12–19. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

