Exploring the plant-associated bacterial communities in Medicago sativa L
- PMID: 22607312
- PMCID: PMC3412730
- DOI: 10.1186/1471-2180-12-78
Exploring the plant-associated bacterial communities in Medicago sativa L
Abstract
Background: Plant-associated bacterial communities caught the attention of several investigators which study the relationships between plants and soil and the potential application of selected bacterial species in crop improvement and protection. Medicago sativa L. is a legume crop of high economic importance as forage in temperate areas and one of the most popular model plants for investigations on the symbiosis with nitrogen fixing rhizobia (mainly belonging to the alphaproteobacterial species Sinorhizobium meliloti). However, despite its importance, no studies have been carried out looking at the total bacterial community associated with the plant. In this work we explored for the first time the total bacterial community associated with M. sativa plants grown in mesocosms conditions, looking at a wide taxonomic spectrum, from the class to the single species (S. meliloti) level.
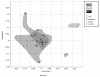
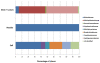
Results: Results, obtained by using Terminal-Restriction Fragment Length Polymorphism (T-RFLP) analysis, quantitative PCR and sequencing of 16 S rRNA gene libraries, showed a high taxonomic diversity as well as a dominance by members of the class Alphaproteobacteria in plant tissues. Within Alphaproteobacteria the families Sphingomonadaceae and Methylobacteriaceae were abundant inside plant tissues, while soil Alphaproteobacteria were represented by the families of Hyphomicrobiaceae, Methylocystaceae, Bradyirhizobiaceae and Caulobacteraceae. At the single species level, we were able to detect the presence of S. meliloti populations in aerial tissues, nodules and soil. An analysis of population diversity on nodules and soil showed a relatively low sharing of haplotypes (30-40%) between the two environments and between replicate mesocosms, suggesting drift as main force shaping S. meliloti population at least in this system.
Conclusions: In this work we shed some light on the bacterial communities associated with M. sativa plants, showing that Alphaproteobacteria may constitute an important part of biodiversity in this system, which includes also the well known symbiont S. meliloti. Interestingly, this last species was also found in plant aerial part, by applying cultivation-independent protocols, and a genetic diversity analysis suggested that population structure could be strongly influenced by random drift.
Figures


References
-
- Mengoni A, Schat H, Vangronsveld J. Plants as extreme environments? Ni-resistant bacteria and Ni-hyperaccumulators of serpentine flora. Plant and Soil. 2010;331:5–16. doi: 10.1007/s11104-009-0242-4. - DOI
-
- Lodewyckx C, Vangronsveld J, Porteous F, Moore ERB, Taghavi S, Mezgeay M, van der Lelie D. Endophytic bacteria and their potential applications. Crit Rev Plant Sci. 2002;21(6):583–606. doi: 10.1080/0735-260291044377. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

