Molecular detection of novel picornaviruses in chickens and turkeys
- PMID: 22160827
- PMCID: PMC7089249
- DOI: 10.1007/s11262-011-0695-4
Molecular detection of novel picornaviruses in chickens and turkeys
Abstract
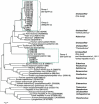
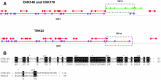
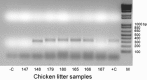
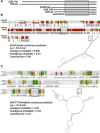
Fecal specimens, including swabs and litter extracts, collected from chickens, domestic ducks, turkeys, and Canadian geese were tested using degenerate primers targeting regions encoding for conserved amino acid motifs (YGDD and DY(T/S)(R/K/G)WDST) in calicivirus RNA-dependent RNA polymerases. Similar motifs are also present in other RNA viruses. Two fecal specimens and 18 litter extracts collected from chickens and turkeys yielded RT-PCR products. BLAST search and phylogenetic analysis revealed that all amplicons represented picornaviruses that clustered into two major groups. Four chicken and one turkey samples yielded 250 bp amplicons with 84-91% nucleotide identity to the recently described turkey hepatitis viruses, while 280 and 283 bp amplicons obtained from 11 chicken and 4 turkey samples represented novel picornaviruses with the closest nucleotide identity to kobuviruses (54-61%) and turdiviruses (47-54%). Analysis of 2.2-3.2 kb extended genome sequences including the partial P2 (2C) and complete P3 (3A, 3B (VPg), 3C(pro), and 3D(pol)) regions of selected strains indicated that viruses yielding the 280/283 bp amplicons represent a putative new genus of Picornaviridae. The 3'-non-translated region (NTR) of the turkey hepatitis-like viruses described in this study was significantly longer (641-654 nt) than that of any of the other piconaviruses and included a putative short open reading frame (ORF). In summary, we report the molecular detection of novel picornaviruses that appear to be endemic in both chickens and turkeys.
Figures


References
-
- N.J. Knowles, T. Hovi, T. Hyypiä, A.M.Q. King, M. Lindberg, M.A. Pallansch, A.C. Palmenberg, P. Simmonds, T. Skern, G. Stanway, T. Yamashita, R. Zell, in Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses, ed. by A.M.Q. King, M.J. Adams, E.B. Carstens, E.J. Lefkowitz (Elsevier, San Diego, 2011), pp. 855–880
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

