comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence
- PMID: 21441318
- PMCID: PMC3126449
- DOI: 10.1128/AEM.00546-11
comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence
Erratum in
- Appl Environ Microbiol. 2011 Jul;77(14):5064
Abstract
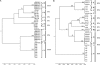
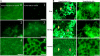
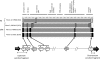
Different strains of Listeria monocytogenes are well known to persist in individual food processing plants and to contaminate foods for many years; however, the specific genotypic and phenotypic mechanisms responsible for persistence of these unique strains remain largely unknown. Based on sequences in comK prophage junction fragments, different strains of epidemic clones (ECs), which included ECII, ECIII, and ECV, were identified and shown to be specific to individual meat and poultry processing plants. The comK prophage-containing strains showed significantly higher cell densities after incubation at 30°C for 48 h on meat and poultry food-conditioning films than did strains lacking the comK prophage (P < 0.05). Overall, the type of strain, the type of conditioning film, and the interaction between the two were all highly significant (P < 0.001). Recombination analysis indicated that the comK prophage junction fragments in these strains had evolved due to extensive recombination. Based on the results of the present study, we propose a novel model in which the concept of defective comK prophage was replaced with the rapid adaptation island (RAI). Genes within the RAI were recharacterized as "adaptons," as these genes may allow L. monocytogenes to rapidly adapt to different food processing facilities and foods. If confirmed, the model presented would help explain Listeria's rapid niche adaptation, biofilm formation, persistence, and subsequent transmission to foods. Also, comK prophage junction fragment sequences may permit accurate tracking of persistent strains back to and within individual food processing operations and thus allow the design of more effective intervention strategies to reduce contamination and enhance food safety.
Figures



References
-
- . 17 April 2009, posting date. Lessons learned: the Canadian Food Inspection Agency's review of Est. 97B (Maple Leaf Consumer Foods Inc.). Canadian Food Inspection Agency, Ottawa, Ontario, Canada
-
- Autio T., Keto-Timonen R., Lunden J., Bjorkroth J., Korkeala H. 2003. Characterisation of persistent and sporadic Listeria monocytogenes strains by pulsed-field gel electrophoresis (PFGE) and amplified fragment length polymorphism (AFLP). Syst. Appl. Microbiol. 26:539–545 - PubMed
-
- Autio T., et al. 2002. Similar Listeria monocytogenes pulsotypes detected in several foods originating from different sources. Int. J. Food Microbiol. 77:83–90 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

