Avian coronavirus in wild aquatic birds
- PMID: 21957308
- PMCID: PMC3209365
- DOI: 10.1128/JVI.05838-11
Avian coronavirus in wild aquatic birds
Abstract
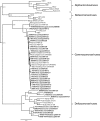
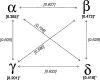
We detected a high prevalence (12.5%) of novel avian coronaviruses in aquatic wild birds. Phylogenetic analyses of these coronaviruses suggest that there is a diversity of gammacoronaviruses and deltacoronaviruses circulating in birds. Gammacoronaviruses were found predominantly in Anseriformes birds, whereas deltacoronaviruses could be detected in Ciconiiformes, Pelecaniformes, and Anseriformes birds in this study. We observed that there are frequent interspecies transmissions of gammacoronaviruses between duck species. In contrast, deltacoronaviruses may have more stringent host specificities. Our analysis of these avian viral and host mitochondrial DNA sequences also suggests that some, but not all, coronaviruses may have coevolved with birds from the same order.
Figures

References
-
- Altizer S., Bartel R., Han B. A. 2011. Animal migration and infectious disease risk. Science 331:296–302 - PubMed
-
- Cavanagh D. 2007. Coronavirus avian infectious bronchitis virus. Vet. Res. 38:281–297 - PubMed
-
- Cavanagh D. 2005. Coronaviruses in poultry and other birds. Avian Pathol. 34:439–448 - PubMed
-
- Chu D. K., et al. 2006. Coronaviruses in bent-winged bats (Miniopterus spp.). J. Gen. Virol. 87:2461–2466 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

