Role of multiple hosts in the cross-species transmission and emergence of a pandemic parvovirus
- PMID: 22072763
- PMCID: PMC3255849
- DOI: 10.1128/JVI.06187-11
Role of multiple hosts in the cross-species transmission and emergence of a pandemic parvovirus
Abstract
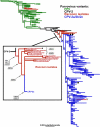
Understanding the mechanisms of cross-species virus transmission is critical to anticipating emerging infectious diseases. Canine parvovirus type 2 (CPV-2) emerged as a variant of a feline parvovirus when it acquired mutations that allowed binding to the canine transferrin receptor type 1 (TfR). However, CPV-2 was soon replaced by a variant virus (CPV-2a) that differed in antigenicity and receptor binding. Here we show that the emergence of CPV involved an additional host range variant virus that has circulated undetected in raccoons for at least 24 years, with transfers to and from dogs. Raccoon virus capsids showed little binding to the canine TfR, showed little infection of canine cells, and had altered antigenic structures. Remarkably, in capsid protein (VP2) phylogenies, most raccoon viruses fell as evolutionary intermediates between the CPV-2 and CPV-2a strains, suggesting that passage through raccoons assisted in the evolution of CPV-2a. This highlights the potential role of alternative hosts in viral emergence.
Figures





References
-
- Agbandje M, McKenna R, Rossmann MG, Strassheim ML, Parrish CR. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155–171 - PubMed
-
- Barros de Freitas R, et al. 2007. The “pressure pan” evolution of human erythrovirus B19 in the Amazon, Brazil. Virology 369:281–287 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

