A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus
- PMID: 22353370
- PMCID: PMC3315249
- DOI: 10.1105/tpc.112.095984
A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus
Abstract
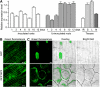
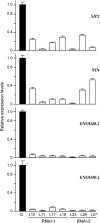
The symbiosis receptor kinase, SymRK, is required for root nodule development. A SymRK-interacting protein (SIP2) was found to form protein complex with SymRK in vitro and in planta. The interaction between SymRK and SIP2 is conserved in legumes. The SIP2 gene was expressed in all Lotus japonicus tissues examined. SIP2 represents a typical plant mitogen-activated protein kinase kinase (MAPKK) and exhibited autophosphorylation and transphosphorylation activities. Recombinant SIP2 protein could phosphorylate casein and the Arabidopsis thaliana MAP kinase MPK6. SymRK and SIP2 could not use one another as a substrate for phosphorylation. Instead, SymRK acted as an inhibitor of SIP2 kinase when MPK6 was used as a substrate, suggesting that SymRK may serve as a negative regulator of the SIP2 signaling pathway. Knockdown expression of SIP2 via RNA interference (RNAi) resulted in drastic reduction of nodules formed in transgenic hairy roots. A significant portion of SIP2 RNAi hairy roots failed to form a nodule. In these roots, the expression levels of SIP2 and three marker genes for infection thread and nodule primordium formation were downregulated drastically, while the expression of two other MAPKK genes were not altered. These observations demonstrate an essential role of SIP2 in the early symbiosis signaling and nodule organogenesis.
Figures




References
-
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 - PubMed
-
- Bardwell L., Cook J.G., Chang E.C., Cairns B.R., Thorner J. (1996). Signaling in the yeast pheromone response pathway: Specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16: 3637–3650 - PMC - PubMed
-
- Beck M., Komis G., Ziemann A., Menzel D., Samaj J. (2011). Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 189: 1069–1083 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

