Functional diversity within the simple gut microbiota of the honey bee
- PMID: 22711827
- PMCID: PMC3390884
- DOI: 10.1073/pnas.1202970109
Functional diversity within the simple gut microbiota of the honey bee
Abstract
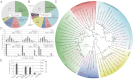
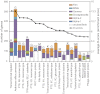
Animals living in social communities typically harbor a characteristic gut microbiota important for nutrition and pathogen defense. Accordingly, in the gut of the honey bee, Apis mellifera, a distinctive microbial community, composed of a taxonomically restricted set of species specific to social bees, has been identified. Despite the ecological and economical importance of honey bees and the increasing concern about population declines, the role of their gut symbionts for colony health and nutrition is unknown. Here, we sequenced the metagenome of the gut microbiota of honey bees. Unexpectedly, we found a remarkable degree of genetic diversity within the few bacterial species colonizing the bee gut. Comparative analysis of gene contents suggests that different species harbor distinct functional capabilities linked to host interaction, biofilm formation, and carbohydrate breakdown. Whereas the former two functions could be critical for pathogen defense and immunity, the latter one might assist nutrient utilization. In a γ-proteobacterial species, we identified genes encoding pectin-degrading enzymes likely involved in the breakdown of pollen walls. Experimental investigation showed that this activity is restricted to a subset of strains of this species providing evidence for niche specialization. Long-standing association of these gut symbionts with their hosts, favored by the eusocial lifestyle of honey bees, might have promoted the genetic and functional diversification of these bee-specific bacteria. Besides revealing insights into mutualistic functions governed by the microbiota of this important pollinator, our findings indicate that the honey bee can serve as a model for understanding more complex gut-associated microbial communities.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Aizen MA, Garibaldi LA, Cunningham SA, Klein AM. Long-term global trends in crop yield and production reveal no current pollination shortage but increasing pollinator dependency. Curr Biol. 2008;18:1572–1575. - PubMed
-
- Evans JD, Schwarz RS. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011;19:614–620. - PubMed
-
- Genersch E. Honey bee pathology: Current threats to honey bees and beekeeping. Appl Microbiol Biotechnol. 2010;87:87–97. - PubMed
-
- Seeley TD. Honeybee Ecology: A Study of Adaptation in Social Life. Princeton: Princeton Univ Press; 1985.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

