Diversifying selection underlies the origin of allozyme polymorphism at the phosphoglucose isomerase locus in Tigriopus californicus
- PMID: 22768211
- PMCID: PMC3386920
- DOI: 10.1371/journal.pone.0040035
Diversifying selection underlies the origin of allozyme polymorphism at the phosphoglucose isomerase locus in Tigriopus californicus
Abstract
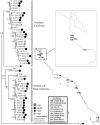
The marine copepod Tigriopus californicus lives in intertidal rock pools along the Pacific coast, where it exhibits strong, temporally stable population genetic structure. Previous allozyme surveys have found high frequency private alleles among neighboring subpopulations, indicating that there is limited genetic exchange between populations. Here we evaluate the factors responsible for the diversification and maintenance of alleles at the phosphoglucose isomerase (Pgi) locus by evaluating patterns of nucleotide variation underlying previously identified allozyme polymorphism. Copepods were sampled from eleven sites throughout California and Baja California, revealing deep genetic structure among populations as well as genetic variability within populations. Evidence of recombination is limited to the sample from Pescadero and there is no support for linkage disequilibrium across the Pgi locus. Neutrality tests and codon-based models of substitution suggest the action of natural selection due to elevated non-synonymous substitutions at a small number of sites in Pgi. Two sites are identified as the charge-changing residues underlying allozyme polymorphisms in T. californicus. A reanalysis of allozyme variation at several focal populations, spanning a period of 26 years and over 200 generations, shows that Pgi alleles are maintained without notable frequency changes. Our data suggest that diversifying selection accounted for the origin of Pgi allozymes, while McDonald-Kreitman tests and the temporal stability of private allozyme alleles suggests that balancing selection may be involved in the maintenance of amino acid polymorphisms within populations.
Conflict of interest statement
Figures
References
-
- McKay JK, Latta RG. Adaptive population divergence: markers, QTL and traits. Trends Ecol Evol. 2002;17:285–291.
-
- Fraser D, Bernatchez L. Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol. 2001;10:2741–2752. - PubMed
-
- Morin PA, Luikart G, Wayne RK the SNP workshop group. SNPs in ecology, evolution and conservation. Trends Ecol Evol. 2004;19:208–216.
-
- Ellegren H, Sheldon BC. Genetic basis of fitness differences in natural populations. Nature. 2008;452:169–175. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

