Human rhinoviruses and enteroviruses in influenza-like illness in Latin America
- PMID: 24119298
- PMCID: PMC3854537
- DOI: 10.1186/1743-422X-10-305
Human rhinoviruses and enteroviruses in influenza-like illness in Latin America
Abstract
Background: Human rhinoviruses (HRVs) belong to the Picornaviridae family with high similarity to human enteroviruses (HEVs). Limited data is available from Latin America regarding the clinical presentation and strains of these viruses in respiratory disease.
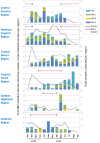
Methods: We collected nasopharyngeal swabs at clinics located in eight Latin American countries from 3,375 subjects aged 25 years or younger who presented with influenza-like illness.
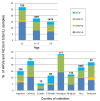
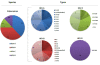
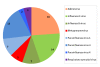
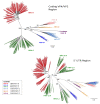
Results: Our subjects had a median age of 3 years and a 1.2:1.0 male:female ratio. HRV was identified in 16% and HEV was identified in 3%. HRVs accounted for a higher frequency of isolates in those of younger age, in particular children < 1 years old. HRV-C accounted for 38% of all HRVs detected. Phylogenetic analysis revealed a high proportion of recombinant strains between HRV-A/HRV-C and between HEV-A/HEV-B. In addition, both EV-D68 and EV-A71 were identified.
Conclusions: In Latin America as in other regions, HRVs and HEVs account for a substantial proportion of respiratory viruses identified in young people with ILI, a finding that provides additional support for the development of pharmaceuticals and vaccines targeting these pathogens.
Figures

References
-
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY. et al.Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;10:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

