A haplotype at STAT2 Introgressed from neanderthals and serves as a candidate of positive selection in Papua New Guinea
- PMID: 22883142
- PMCID: PMC3415544
- DOI: 10.1016/j.ajhg.2012.06.015
A haplotype at STAT2 Introgressed from neanderthals and serves as a candidate of positive selection in Papua New Guinea
Abstract
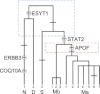
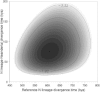
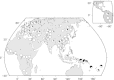
Signals of archaic admixture have been identified through comparisons of the draft Neanderthal and Denisova genomes with those of living humans. Studies of individual loci contributing to these genome-wide average signals are required for characterization of the introgression process and investigation of whether archaic variants conferred an adaptive advantage to the ancestors of contemporary human populations. However, no definitive case of adaptive introgression has yet been described. Here we provide a DNA sequence analysis of the innate immune gene STAT2 and show that a haplotype carried by many Eurasians (but not sub-Saharan Africans) has a sequence that closely matches that of the Neanderthal STAT2. This haplotype, referred to as N, was discovered through a resequencing survey of the entire coding region of STAT2 in a global sample of 90 individuals. Analyses of publicly available complete genome sequence data show that haplotype N shares a recent common ancestor with the Neanderthal sequence (~80 thousand years ago) and is found throughout Eurasia at an average frequency of ~5%. Interestingly, N is found in Melanesian populations at ~10-fold higher frequency (~54%) than in Eurasian populations. A neutrality test that controls for demography rejects the hypothesis that a variant of N rose to high frequency in Melanesia by genetic drift alone. Although we are not able to pinpoint the precise target of positive selection, we identify nonsynonymous mutations in ERBB3, ESYT1, and STAT2-all of which are part of the same 250 kb introgressive haplotype-as good candidates.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Hawks J., Cochran G., Harpending H.C., Lahn B.T. A genetic legacy from archaic Homo. Trends Genet. 2008;24:19–23. - PubMed
-
- Hardy J., Pittman A., Myers A., Gwinn-Hardy K., Fung H.C., de Silva R., Hutton M., Duckworth J. Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens. Biochem. Soc. Trans. 2005;33:582–585. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

