Next generation sequencing in predicting gene function in podophyllotoxin biosynthesis
- PMID: 23161544
- PMCID: PMC3537044
- DOI: 10.1074/jbc.M112.400689
Next generation sequencing in predicting gene function in podophyllotoxin biosynthesis
Abstract
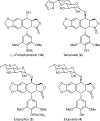
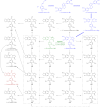
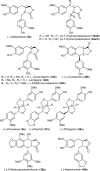
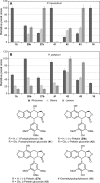
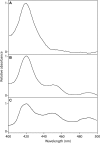
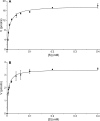
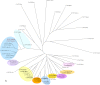
Podophyllum species are sources of (-)-podophyllotoxin, an aryltetralin lignan used for semi-synthesis of various powerful and extensively employed cancer-treating drugs. Its biosynthetic pathway, however, remains largely unknown, with the last unequivocally demonstrated intermediate being (-)-matairesinol. Herein, massively parallel sequencing of Podophyllum hexandrum and Podophyllum peltatum transcriptomes and subsequent bioinformatics analyses of the corresponding assemblies were carried out. Validation of the assembly process was first achieved through confirmation of assembled sequences with those of various genes previously established as involved in podophyllotoxin biosynthesis as well as other candidate biosynthetic pathway genes. This contribution describes characterization of two of the latter, namely the cytochrome P450s, CYP719A23 from P. hexandrum and CYP719A24 from P. peltatum. Both enzymes were capable of converting (-)-matairesinol into (-)-pluviatolide by catalyzing methylenedioxy bridge formation and did not act on other possible substrates tested. Interestingly, the enzymes described herein were highly similar to methylenedioxy bridge-forming enzymes from alkaloid biosynthesis, whereas candidates more similar to lignan biosynthetic enzymes were catalytically inactive with the substrates employed. This overall strategy has thus enabled facile further identification of enzymes putatively involved in (-)-podophyllotoxin biosynthesis and underscores the deductive power of next generation sequencing and bioinformatics to probe and deduce medicinal plant biosynthetic pathways.
Figures



References
-
- Ansorge W. J. (2009) Next-generation DNA sequencing techniques. N. Biotechnol. 25, 195–203 - PubMed
-
- Chen S., Luo H., Li Y., Sun Y., Wu Q., Niu Y., Song J., Lv A., Zhu Y., Sun C., Steinmetz A., Qian Z. (2011) 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Rep. 30, 1593–1601 - PubMed
-
- Subramaniyam S., Mathiyalagan R., Jun Gyo I., Bum-Soo L., Sungyoung L., Deok Chun Y. (2011) Transcriptome profiling and in silico analysis of Gynostemma pentaphyllum using a next generation sequencer. Plant Cell Rep. 30, 2075–2083 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

