Diverse broad-host-range plasmids from freshwater carry few accessory genes
- PMID: 24096417
- PMCID: PMC3837812
- DOI: 10.1128/AEM.02252-13
Diverse broad-host-range plasmids from freshwater carry few accessory genes
Abstract
Broad-host-range self-transferable plasmids are known to facilitate bacterial adaptation by spreading genes between phylogenetically distinct hosts. These plasmids typically have a conserved backbone region and a variable accessory region that encodes host-beneficial traits. We do not know, however, how well plasmids that do not encode accessory functions can survive in nature. The goal of this study was to characterize the backbone and accessory gene content of plasmids that were captured from freshwater sources without selecting for a particular phenotype or cultivating their host. To do this, triparental matings were used such that the only required phenotype was the plasmid's ability to mobilize a nonconjugative plasmid. Based on complete genome sequences of 10 plasmids, only 5 carried identifiable accessory gene regions, and none carried antibiotic resistance genes. The plasmids belong to four known incompatibility groups (IncN, IncP-1, IncU, and IncW) and two potentially new groups. Eight of the plasmids were shown to have a broad host range, being able to transfer into alpha-, beta-, and gammaproteobacteria. Because of the absence of antibiotic resistance genes, we resampled one of the sites and compared the proportion of captured plasmids that conferred antibiotic resistance to their hosts with the proportion of such plasmids captured from the effluent of a local wastewater treatment plant. Few of the captured plasmids from either site encoded antibiotic resistance. A high diversity of plasmids that encode no or unknown accessory functions is thus readily found in freshwater habitats. The question remains how the plasmids persist in these microbial communities.
Figures


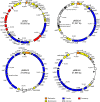
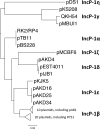
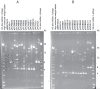
References
-
- Thomas CM. 2004. Evolution and population genetics of bacterial plasmids p 509–528 In Funnell BE, Phillips GJ. (ed), Plasmid biology. ASM Press, Washington, DC
-
- Szpirer C, Top E, Couturier M, Mergeay M. 1999. Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology 145:3321–3329 - PubMed
-
- Datta N, Hedges RW. 1972. Host ranges of R factors. J. Gen. Microbiol. 70:453–460 - PubMed
-
- Thomas CM, Smith CA. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 41:77–101 - PubMed
-
- Fernandez-Lopez R, Garcillan-Barcia M, Revilla C, Lazaro M, Vielva L, de la Cruz F. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942–966 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

