Twelve previously unknown phage genera are ubiquitous in global oceans
- PMID: 23858439
- PMCID: PMC3732932
- DOI: 10.1073/pnas.1305956110
Twelve previously unknown phage genera are ubiquitous in global oceans
Abstract
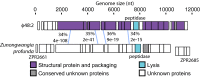
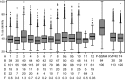
Viruses are fundamental to ecosystems ranging from oceans to humans, yet our ability to study them is bottlenecked by the lack of ecologically relevant isolates, resulting in "unknowns" dominating culture-independent surveys. Here we present genomes from 31 phages infecting multiple strains of the aquatic bacterium Cellulophaga baltica (Bacteroidetes) to provide data for an underrepresented and environmentally abundant bacterial lineage. Comparative genomics delineated 12 phage groups that (i) each represent a new genus, and (ii) represent one novel and four well-known viral families. This diversity contrasts the few well-studied marine phage systems, but parallels the diversity of phages infecting human-associated bacteria. Although all 12 Cellulophaga phages represent new genera, the podoviruses and icosahedral, nontailed ssDNA phages were exceptional, with genomes up to twice as large as those previously observed for each phage type. Structural novelty was also substantial, requiring experimental phage proteomics to identify 83% of the structural proteins. The presence of uncommon nucleotide metabolism genes in four genera likely underscores the importance of scavenging nutrient-rich molecules as previously seen for phages in marine environments. Metagenomic recruitment analyses suggest that these particular Cellulophaga phages are rare and may represent a first glimpse into the phage side of the rare biosphere. However, these analyses also revealed that these phage genera are widespread, occurring in 94% of 137 investigated metagenomes. Together, this diverse and novel collection of phages identifies a small but ubiquitous fraction of unknown marine viral diversity and provides numerous environmentally relevant phage-host systems for experimental hypothesis testing.
Keywords: model systems; phage genomics; phage taxonomy; prophage; queuosine biosynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320(5879):1034–1039. - PubMed
-
- Breitbart M. Marine viruses: Truth or dare. Annu Rev Mar Sci. 2012;4:5.1–5.24. - PubMed
-
- Borriss M, et al. Genome and proteome characterization of the psychrophilic Flavobacterium bacteriophage 11b. Extremophiles. 2007;11(1):95–104. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

