Evaluation of four single-locus markers for Leishmania species discrimination by sequencing
- PMID: 24452158
- PMCID: PMC3993476
- DOI: 10.1128/JCM.02936-13
Evaluation of four single-locus markers for Leishmania species discrimination by sequencing
Abstract
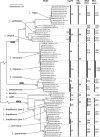
Several genetic markers have been described for discriminating Leishmania species. In most reported cases, one or a few polymorphisms are the basis of species identification, and the methods were validated on a limited number of strains from a particular geographical region. Therefore, most techniques may underestimate the global intraspecies variability and are applicable only in certain areas. In addition, interlaboratory standardization is mostly absent, complicating comparisons among different studies. Here, we compared species typing results from all sequence polymorphisms found in four popular markers that can be applied directly on clinical samples: the miniexon or spliced leader, the internal transcribed spacer of the ribosomal DNA array, the 7SL RNA gene, and the heat shock protein 70 gene. Clustering was evaluated among 74 Leishmania strains, selected to represent a wide geographic distribution and genetic variability of the medically relevant species of the genus. Results were compared with a multilocus sequence typing (MLST) approach using 7 single-copy household genes and with multilocus enzyme electrophoresis (MLEE), still considered the gold standard by some. We show that strain groupings are highly congruent across the four different single-locus markers, MLST, and MLEE. Overall, the heat shock protein 70 gene and the miniexon presented the best resolutions for separating medically relevant species. As gene sequence analysis is validated here on a global scale, it is advocated as the method of choice for use in genetic, clinical, and epidemiological studies and for managing patients with unknown origins of infection, especially in Western infectious disease clinics dealing with imported leishmaniasis.
Figures

References
-
- Romero GA, Guerra MV, Paes MG, Macedo VO. 2001. Comparison of cutaneous leishmaniasis due to Leishmania (Viannia) braziliensis and L. (V.) guyanensis in Brazil: therapeutic response to meglumine antimoniate. Am. J. Trop. Med. Hyg. 65:456–465 - PubMed
-
- Arevalo J, Ramirez L, Adaui V, Zimic M, Tulliano G, Miranda-Verastegui C, Lazo M, Loayza-Muro R, De Doncker S, Maurer A, Chappuis F, Dujardin JC, Llanos-Cuentas A. 2007. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J. Infect. Dis. 195:1846–1851. 10.1086/518041 - DOI - PubMed
-
- Marfurt J, Nasereddin A, Niederwieser I, Jaffe CL, Beck HP, Felger I. 2003. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J. Clin. Microbiol. 41:3147–3153. 10.1128/JCM.41.7.3147-3153.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

