Local phylogenetic analysis identifies distinct trends in transmitted HIV drug resistance: implications for public health interventions
- PMID: 24171696
- PMCID: PMC3816547
- DOI: 10.1186/1471-2334-13-509
Local phylogenetic analysis identifies distinct trends in transmitted HIV drug resistance: implications for public health interventions
Abstract
Background: HIV transmitted drug resistance (TDR) surveillance is usually conducted by sampling from a large population. However, overall TDR prevalence results may be inaccurate for many individual clinical setting. We analyzed HIV genotypes at a tertiary care setting in Ottawa, Ontario in order to evaluate local TDR patterns among sub-populations.
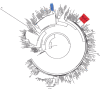
Method: Genotyping reports were digitized from ART naïve patients followed at the Immunodeficiency Clinic at the Ottawa Hospital, between 2008 and 2010. Quality controlled, digitized sequence data were assessed for TDR using the Stanford HIV Database. Patient characteristics were analyzed according to TDR patterns. Finally, a phylogenetic tree was constructed to elucidate the observed pattern of HIV TDR.
Results: Among the 155 clinic patients there was no statistically significantly difference in demographics as compared to the Ontario provincial HIV population. The clinic prevalence of TDR was 12.3%; however, in contrast to the data from Ontario, TDR patterns were inverted with a 21% prevalence among MSM and 5.5% among IDU. Furthermore, nearly 80% of the observed TDR was a D67N/K219Q pattern with 87% of these infections arising from a distinct phylogenetic cluster.
Conclusions: Local patterns of TDR were distinct to what had been observed provincially. Phylogenetic analysis uncovered a cluster of related infections among MSM that appeared more likely to be recent infections. Results support a paradigm of routine local TDR surveillance to identify the sub-populations under care. Furthermore, the routine application of phylogenetic analysis in the TDR surveillance context provides insights into how best to target prevention strategies; and how to correctly measure outcomes.
Figures
References
-
- Palella FJ, Armon C, Buchacz K, Cole SR, Chmiel JS, Novak RM, Wood K, Moorman AC, Brooks JT. HOPS (HIV Outpatient Study) Investigators: The association of HIV susceptibility testing with survival among HIV-infected patients receiving antiretroviral therapy: a cohort study. Ann Intern Med. 2009;151:73–84. doi: 10.7326/0003-4819-151-2-200907210-00003. - DOI - PubMed
-
- DeGruttola V, Dix L, D’Aquila R, Holder D, Phillips A, Ait-Khaled M, Baxter J, Clevenbergh P, Hammer S, Harrigan R, Katzenstein D, Lanier R, Miller M, Para M, Yerly S, Zolopa A, Murray J, Patick A, Miller V, Castillo S, Pedneault L, Mellors J. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antiver Ther. 2000;5:41–48. - PubMed
-
- Méchai F, Quertainmont Y, Sahali S, Delfraissy J-F, Ghosn J. Post-exposure prophylaxis with a maraviroc-containing regimen after occupational exposure to a multi-resistant HIV-infected source person. J Med Virol. 2007;80:9–10. - PubMed
-
- Vercauteren J, Wensing AMJ, van de Vijver DAMC, Albert J, Balotta C, Hamouda O, Kucherer C, Struck D, Schmit J-C, Asjö B, Bruckova M, Camacho RJ, Clotet B, Coughlan S, Grossman Z, Horban A, Korn K, Kostrikis L, Nielsen C, Paraskevis D, Poljak M, Puchhammer-Stöckl E, Riva C, Ruiz L, Salminen M, Schuurman R, Sonnerborg A, Stanekova D, Stanojevic M, Vandamme A-M, Boucher CAB. Transmission of Drug‒Resistant HIV‒1 Is Stabilizing in Europe. J Infect Dis. 2009;200:1503–1508. doi: 10.1086/644505. - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

