Phylodynamics of vampire bat-transmitted rabies in Argentina
- PMID: 24661865
- PMCID: PMC4870601
- DOI: 10.1111/mec.12728
Phylodynamics of vampire bat-transmitted rabies in Argentina
Abstract
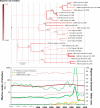
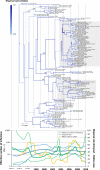
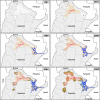
Common vampire bat populations distributed from Mexico to Argentina are important rabies reservoir hosts in Latin America. The aim of this work was to analyse the population structure of the rabies virus (RABV) variants associated with vampire bats in the Americas and to study their phylodynamic pattern within Argentina. The phylogenetic analysis based on all available vampire bat-related N gene sequences showed both a geographical and a temporal structure. The two largest groups of RABV variants from Argentina were isolated from northwestern Argentina and from the central western zone of northeastern Argentina, corresponding to livestock areas with different climatic, topographic and biogeographical conditions, which determined their dissemination and evolutionary patterns. In addition, multiple introductions of the infection into Argentina, possibly from Brazil, were detected. The phylodynamic analysis suggests that RABV transmission dynamics is characterized by initial epizootic waves followed by local enzootic cycles with variable persistence. Anthropogenic interventions in the ecosystem should be assessed taking into account not only the environmental impact but also the potential risk of disease spreading through dissemination of current RABV lineages or the emergence of novel ones associated with vampire bats.
Keywords: Argentina; phylodynamics; phylogeography; rabies virus; vampire bats.
© 2014 John Wiley & Sons Ltd.
Figures


References
-
- Barbosa TF, Medeiros DB, Travassos da Rosa ES, et al. Molecular epidemiology of rabies virus isolated from different sources during a bat-transmitted human outbreak occurring in Augusto Correa municipality, Brazilian Amazon. Virology. 2008;370:228–236. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

