An ancestral bacterial division system is widespread in eukaryotic mitochondria
- PMID: 25831547
- PMCID: PMC4547283
- DOI: 10.1073/pnas.1421392112
An ancestral bacterial division system is widespread in eukaryotic mitochondria
Abstract
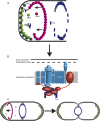
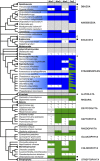
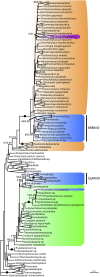
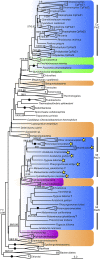
Bacterial division initiates at the site of a contractile Z-ring composed of polymerized FtsZ. The location of the Z-ring in the cell is controlled by a system of three mutually antagonistic proteins, MinC, MinD, and MinE. Plastid division is also known to be dependent on homologs of these proteins, derived from the ancestral cyanobacterial endosymbiont that gave rise to plastids. In contrast, the mitochondria of model systems such as Saccharomyces cerevisiae, mammals, and Arabidopsis thaliana seem to have replaced the ancestral α-proteobacterial Min-based division machinery with host-derived dynamin-related proteins that form outer contractile rings. Here, we show that the mitochondrial division system of these model organisms is the exception, rather than the rule, for eukaryotes. We describe endosymbiont-derived, bacterial-like division systems comprising FtsZ and Min proteins in diverse less-studied eukaryote protistan lineages, including jakobid and heterolobosean excavates, a malawimonad, stramenopiles, amoebozoans, a breviate, and an apusomonad. For two of these taxa, the amoebozoan Dictyostelium purpureum and the jakobid Andalucia incarcerata, we confirm a mitochondrial localization of these proteins by their heterologous expression in Saccharomyces cerevisiae. The discovery of a proteobacterial-like division system in mitochondria of diverse eukaryotic lineages suggests that it was the ancestral feature of all eukaryotic mitochondria and has been supplanted by a host-derived system multiple times in distinct eukaryote lineages.
Keywords: Min proteins; MinCDE; mitochondria; mitochondrial division; mitochondrial fission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Meier EL, Goley ED. Form and function of the bacterial cytokinetic ring. Curr Opin Cell Biol. 2014;26:19–27. - PubMed
-
- Natale P, Pazos M, Vicente M. The Escherichia coli divisome: Born to divide. Environ Microbiol. 2013;15(12):3169–3182. - PubMed
-
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

