Genetic analysis of members of the species Oropouche virus and identification of a novel M segment sequence
- PMID: 25735305
- PMCID: PMC4635451
- DOI: 10.1099/vir.0.000108
Genetic analysis of members of the species Oropouche virus and identification of a novel M segment sequence
Abstract
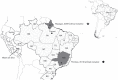
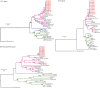
Oropouche virus (OROV) is a public health threat in South America, and in particular in northern Brazil, causing frequent outbreaks of febrile illness. Using a combination of deep sequencing and Sanger sequencing approaches, we determined the complete genome sequences of eight clinical isolates that were obtained from patient sera during an Oropouche fever outbreak in Amapa state, northern Brazil, in 2009. We also report the complete genome sequences of two OROV reassortants isolatd from two marmosets in Minas Gerais state, south-east Brazil, in 2012 that contained a novel M genome segment. Interestingly, all 10 isolates possessed a 947 nt S segment that lacked 11 residues in the S-segment 3' UTR compared with the recently redetermined Brazilian prototype OROV strain BeAn19991. OROV maybe circulating more widely in Brazil and in the non-human primate population than previously appreciated, and the identification of yet another reassortant highlights the importance of bunyavirus surveillance in South America.
Figures



References
-
- Acrani G. O., Tilston-Lunel N. L., Spiegel M., Weidemann M., Dilcher M., Andrade da Silva D. E., Nunes M. R. T., Elliott R. M. ( 2014. ). Establishment of a minigenome system for Oropouche virus reveals the S genome segment to be significantly longer than reported previously. J Gen Virol 96, 513–523.10.1099/jgv.0.000005 - DOI - PubMed
-
- Aguilar P. V., Barrett A. D., Saeed M. F., Watts D. M., Russell K., Guevara C., Ampuero J. S., Suarez L., Cespedes M., et al. ( 2011. ). Iquitos virus: a novel reassortant Orthobunyavirus associated with human illness in Peru. PLoS Negl Trop Dis 5, e1315. 10.1371/journal.pntd.0001315 - DOI - PMC - PubMed
-
- Anderson C. R., Spence L., Downs W. G., Aitken T. H. ( 1961. ). Oropouche virus: a new human disease agent from Trinidad, West Indies. Am J Trop Med Hyg 10, 574–578. - PubMed
-
- Baisley K. J., Watts D. M., Munstermann L. E., Wilson M. L. ( 1998. ). Epidemiology of endemic Oropouche virus transmission in upper Amazonian Peru. Am J Trop Med Hyg 59, 710–716. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

