Successive Respiratory Syncytial Virus Epidemics in Local Populations Arise from Multiple Variant Introductions, Providing Insights into Virus Persistence
- PMID: 26355091
- PMCID: PMC4645665
- DOI: 10.1128/JVI.01972-15
Successive Respiratory Syncytial Virus Epidemics in Local Populations Arise from Multiple Variant Introductions, Providing Insights into Virus Persistence
Abstract
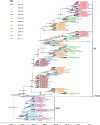
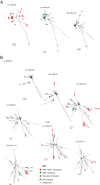
Respiratory syncytial virus (RSV) is a global respiratory pathogen of humans, with infection occurring characteristically as recurrent seasonal epidemics. Unlike influenza viruses, little attention has been paid to the mechanism underlying worldwide spread and persistence of RSV and how this may be discerned through an improved understanding of the introduction and persistence of RSV in local communities. We analyzed 651 attachment (G) glycoprotein nucleotide sequences of RSV B collected over 11 epidemics (2002 to 2012) in Kilifi, Kenya, and contemporaneous data collected elsewhere in Kenya and 18 other countries worldwide (2002 to 2012). Based on phylogeny, genetic distance and clustering patterns, we set out pragmatic criteria to classify local viruses into distinct genotypes and variants, identifying those newly introduced and those locally persisting. Three genotypes were identified in the Kilifi data set: BA (n = 500), SAB1 (n = 148), and SAB4 (n = 3). Recurrent RSV epidemics in the local population were composed of numerous genetic variants, most of which have been newly introduced rather than persisting in the location from season to season. Global comparison revealed that (i) most Kilifi variants do not cluster closely with strains from outside Kenya, (ii) some Kilifi variants were closely related to those observed outside Kenya (mostly Western Europe), and (iii) many variants were circulating elsewhere but were never detected in Kilifi. These results are consistent with the hypothesis that year-to-year presence of RSV at the local level (i.e., Kilifi) is achieved primarily, but not exclusively, through introductions from a pool of variants that are geographically restricted (i.e., to Kenya or to the region) rather than global.
Importance: The mechanism by which RSV persists and reinvades local populations is poorly understood. We investigated this by studying the temporal patterns of RSV variants in a rural setting in tropical Africa and comparing these variants with contemporaneous variants circulating in other countries. We found that periodic seasonal RSV transmission at the local level appears to require regular new introductions of variants. However, importantly, the evidence suggests that the source of new variants is mostly geographically restricted, and we hypothesize that year-to-year RSV persistence is at the country level rather than the global level. This has implications for control.
Copyright © 2015, Agoti et al.
Figures




References
-
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi:10.1016/S0140-6736(10)60206-1. - DOI - PMC - PubMed
-
- Haynes AK, Manangan AP, Iwane MK, Sturm-Ramirez K, Homaira N, Brooks WA, Luby S, Rahman M, Klena JD, Zhang Y, Yu H, Zhan F, Dueger E, Mansour AM, Azazzy N, McCracken JP, Bryan JP, Lopez MR, Burton DC, Bigogo G, Breiman RF, Feikin DR, Njenga K, Montgomery J, Cohen AL, Moyes J, Pretorius M, Cohen C, Venter M, Chittaganpitch M, Thamthitiwat S, Sawatwong P, Baggett HC, Luber G, Gerber SI. 2013. Respiratory syncytial virus circulation in seven countries with Global Disease Detection Regional Centers. J Infect Dis 208(Suppl 3):S246–S254. doi:10.1093/infdis/jit515. - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

