Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist
- PMID: 26283367
- PMCID: PMC4568208
- DOI: 10.1073/pnas.1508737112
Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist
Abstract
The trillions of bacteria that make up the human microbiome are believed to encode functions that are important to human health; however, little is known about the specific effectors that commensal bacteria use to interact with the human host. Functional metagenomics provides a systematic means of surveying commensal DNA for genes that encode effector functions. Here, we examine 3,000 Mb of metagenomic DNA cloned from three phenotypically distinct patients for effectors that activate NF-κB, a transcription factor known to play a central role in mediating responses to environmental stimuli. This screen led to the identification of 26 unique commensal bacteria effector genes (Cbegs) that are predicted to encode proteins with diverse catabolic, anabolic, and ligand-binding functions and most frequently interact with either glycans or lipids. Detailed analysis of one effector gene family (Cbeg12) recovered from all three patient libraries found that it encodes for the production of N-acyl-3-hydroxypalmitoyl-glycine (commendamide). This metabolite was also found in culture broth from the commensal bacterium Bacteroides vulgatus, which harbors a gene highly similar to Cbeg12. Commendamide resembles long-chain N-acyl-amides that function as mammalian signaling molecules through activation of G-protein-coupled receptors (GPCRs), which led us to the observation that commendamide activates the GPCR G2A/GPR132. G2A has been implicated in disease models of autoimmunity and atherosclerosis. This study shows the utility of functional metagenomics for identifying potential mechanisms used by commensal bacteria for host interactions and outlines a functional metagenomics-based pipeline for the systematic identification of diverse commensal bacteria effectors that impact host cellular functions.
Keywords: Cbeg; N-acyl amino acids; NF-κB; commendamide; functional metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



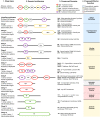
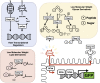


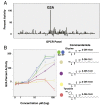
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

