T-cell epitope content comparison (EpiCC) of swine H1 influenza A virus hemagglutinin
- PMID: 29054116
- PMCID: PMC5705686
- DOI: 10.1111/irv.12513
T-cell epitope content comparison (EpiCC) of swine H1 influenza A virus hemagglutinin
Abstract
Background: Predicting vaccine efficacy against emerging pathogen strains is a significant problem in human and animal vaccine design. T-cell epitope cross-conservation may play an important role in cross-strain vaccine efficacy. While influenza A virus (IAV) hemagglutination inhibition (HI) antibody titers are widely used to predict protective efficacy of 1 IAV vaccine against new strains, no similar correlate of protection has been identified for T-cell epitopes.
Objective: We developed a computational method (EpiCC) that facilitates pairwise comparison of protein sequences based on an immunological property-T-cell epitope content-rather than sequence identity, and evaluated its ability to classify swine IAV strain relatedness to estimate cross-protective potential of a vaccine strain for circulating viruses.
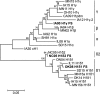
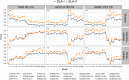
Methods: T-cell epitope relatedness scores were assessed for 23 IAV HA sequences representing the major H1 swine IAV phylo-clusters circulating in North American swine and HA sequences in a commercial inactivated vaccine (FluSure XP® ). Scores were compared to experimental data from previous efficacy studies.
Results: Higher EpiCC scores were associated with greater protection by the vaccine against strains for 23 field IAV strain vaccine comparisons. A threshold for EpiCC relatedness associated with full or partial protection in the absence of cross-reactive HI antibodies was identified. EpiCC scores for field strains for which FluSure protective efficacy is not yet available were also calculated.
Conclusion: EpiCC thresholds can be evaluated for predictive accuracy of protection in future efficacy studies. EpiCC may also complement HI cross-reactivity and phylogeny for selection of influenza strains in vaccine development.
Keywords: T-cell epitope content comparison; T-cell epitope prediction; computational immunology; hemagglutinin; influenza A viruses; swine influenza H1 viruses; swine leukocyte antigen; vaccine efficacy.
© 2017 The Authors. Influenza and Other Respiratory Viruses Published by John Wiley & Sons Ltd.
Figures
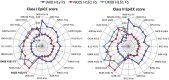
References
-
- Vincent A, Awada L, Brown I, et al. Review of influenza a virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health. 2014;61:4‐17. - PubMed
-
- Anderson TK, Campbell BA, Nelson MI, et al. Characterization of co‐circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus Res. 2015;201:24‐31. - PubMed
-
- Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes. 2009;39:176‐185. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

