Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness
- PMID: 28069935
- PMCID: PMC5278458
- DOI: 10.1073/pnas.1620144114
Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness
Abstract
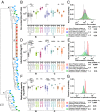
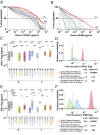
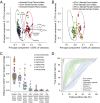
Sexual transmission of HIV-1 is an inefficient process, with only one or few variants of the donor quasispecies establishing the new infection. A critical, and as yet unresolved, question is whether the mucosal bottleneck selects for viruses with increased transmission fitness. Here, we characterized 300 limiting dilution-derived virus isolates from the plasma, and in some instances genital secretions, of eight HIV-1 donor and recipient pairs. Although there were no differences in the amount of virion-associated envelope glycoprotein, recipient isolates were on average threefold more infectious (P = 0.0001), replicated to 1.4-fold higher titers (P = 0.004), were released from infected cells 4.2-fold more efficiently (P < 0.00001), and were significantly more resistant to type I IFNs than the corresponding donor isolates. Remarkably, transmitted viruses exhibited 7.8-fold higher IFNα2 (P < 0.00001) and 39-fold higher IFNβ (P < 0.00001) half-maximal inhibitory concentrations (IC50) than did donor isolates, and their odds of replicating in CD4+ T cells at the highest IFNα2 and IFNβ doses were 35-fold (P < 0.00001) and 250-fold (P < 0.00001) greater, respectively. Interestingly, pretreatment of CD4+ T cells with IFNβ, but not IFNα2, selected donor plasma isolates that exhibited a transmitted virus-like phenotype, and such viruses were also detected in the donor genital tract. These data indicate that transmitted viruses are phenotypically distinct, and that increased IFN resistance represents their most distinguishing property. Thus, the mucosal bottleneck selects for viruses that are able to replicate and spread efficiently in the face of a potent innate immune response.
Keywords: HIV-1 transmission fitness; HIV-1 transmission pairs; innate immunity; mucosal HIV-1 transmission; type I interferons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- UNAIDS 2016. Global AIDS Update 2016. Available at www.unaids.org/sites/default/files/media_asset/global-AIDS-update-2016_e.... Accessed December 21, 2016.
-
- Burgener A, McGowan I, Klatt NR. HIV and mucosal barrier interactions: consequences for transmission and pathogenesis. Curr Opin Immunol. 2015;36:22–30. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

