Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9
- PMID: 25410609
- PMCID: PMC4263139
- DOI: 10.1038/ncomms6560
Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9
Abstract
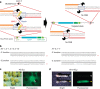
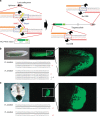
Genome engineering using programmable nucleases enables homologous recombination (HR)-mediated gene knock-in. However, the labour used to construct targeting vectors containing homology arms and difficulties in inducing HR in some cell type and organisms represent technical hurdles for the application of HR-mediated knock-in technology. Here, we introduce an alternative strategy for gene knock-in using transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) mediated by microhomology-mediated end-joining, termed the PITCh (Precise Integration into Target Chromosome) system. TALEN-mediated PITCh, termed TAL-PITCh, enables efficient integration of exogenous donor DNA in human cells and animals, including silkworms and frogs. We further demonstrate that CRISPR/Cas9-mediated PITCh, termed CRIS-PITCh, can be applied in human cells without carrying the plasmid backbone sequence. Thus, our PITCh-ing strategies will be useful for a variety of applications, not only in cultured cells, but also in various organisms, including invertebrates and vertebrates.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

