Dynamic variation of the microbial community structure during the long-time mono-fermentation of maize and sugar beet silage
- PMID: 25712194
- PMCID: PMC4554465
- DOI: 10.1111/1751-7915.12263
Dynamic variation of the microbial community structure during the long-time mono-fermentation of maize and sugar beet silage
Abstract
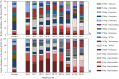
This study investigated the development of the microbial community during a long-term (337 days) anaerobic digestion of maize and sugar beet silage, two feedstocks that significantly differ in their chemical composition. For the characterization of the microbial dynamics, the community profiling method terminal restriction fragment length polymorphism (TRFLP) in combination with a cloning-sequencing approach was applied. Our results revealed a specific adaptation of the microbial community to the supplied feedstocks. Based on the high amount of complex compounds, the anaerobic conversion rate of maize silage was slightly lower compared with the sugar beet silage. It was demonstrated that members from the phylum Bacteroidetes are mainly involved in the degradation of low molecular weight substances such as sugar, ethanol and acetate, the main compounds of the sugar beet silage. It was further shown that species of the genus Methanosaeta are highly sensitive against sudden stress situations such as a strong decrease in the ammonium nitrogen (NH₄(+)-N) concentration or a drop of the pH value. In both cases, a functional compensation by members of the genera Methanoculleus and/or Methanosarcina was detected. However, the overall biomass conversion of both feedstocks proceeded efficiently as a steady state between acid production and consumption was recorded, which further resulted in an equal biogas yield.
© 2015 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures




References
-
- Balussou D, Kleyböcker A, McKenna R, Möst D. Fichtner W. An economic analysis of three operational co-digestion biogas plants in Germany. Waste Biomass Valorization. 2012;3:23–41.
-
- Breure A, Mooijman K. Van Andel J. Protein degradation in anaerobic digestion: influence of volatile fatty acids and carbohydrates on hydrolysis and acidogenic fermentation of gelatin. Appl Microbiol Biotechnol. 1986;24:426–431.
-
- Cai S, Li J, Hu FZ, Zhang K, Luo Y, Janto B, et al. Cellulosilyticum ruminicola, a newly described rumen bacterium that possesses redundant fibrolytic-protein-encoding genes and degrades lignocellulose with multiple carbohydrate-borne fibrolytic enzymes. Appl Environ Microbiol. 2010;76:3818–3824. - PMC - PubMed
-
- Carballa M, Smits M, Etchebehere C, Boon N. Verstraete W. Correlations between molecular and operational parameters in continuous lab-scale anaerobic reactors. Appl Microbiol Biotechnol. 2011;89:303–314. - PubMed
-
- Chen S. Dong X. Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int J Syst Evol Microbiol. 2005;55:2257–2261. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

