Proteogenomic verifies targets underlying erythromycin alleviate neutrophil extracellular traps-induced inflammation
- PMID: 40253327
- PMCID: PMC12009532
- DOI: 10.1186/s12931-025-03226-5
Proteogenomic verifies targets underlying erythromycin alleviate neutrophil extracellular traps-induced inflammation
Abstract
Background: Neutrophil Extracellular Traps (NETs) are closely related to the progression of inflammation in Chronic Obstructive Pulmonary Disease (COPD). Erythromycin (EM) has been shown to inhibit inflammation in COPD, but its molecular mechanisms is still unclear. The aim of our study is investigate the molecular mechanisms of EM's anti-inflammatory effects in NETs-induced inflammation.
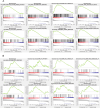
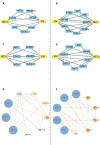
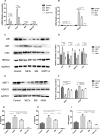
Methods: Transcriptomics and proteomics data were obtained from U937 cells treated with NETs and EM. Differentially expressed genes (DEGs) and differentially expressed proteins (DEPs) were identified using R software. Pathway enrichment analyses, were employed to identify inflammation-related pathways. Cytoscape were utilized to construct network of hub targets regulated by EM which related with oxidative stress and inflammation. Additionally, Cytoscape and STRING were used to construct protein-protein interaction (PPI) network of key targets regulated by EM. The expression levels of key targets were further confirmed through WB and PCR experiments.
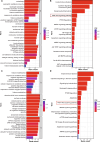
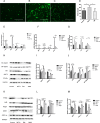
Results: Both transcriptomics and proteomics indicate that EM decrease NETs -induced AKT1 expression. Enrichment analysis of DEGs and DEPs reveal multiple common pathways involved in EM's regulation inflammation, including the PI3K/AKT pathway, response to oxidative stress, IKK/NF-κB signaling and PTEN signaling pathway. Nine key targets in PI3K/AKT-related inflammatory pathways regulated by EM and ten targets of EM-regulated oxidative stress were identified. WB and PCR results confirmed that EM reversing the NETs-induced inflammation by modulating the activity of these targets. Furthermore, clinical samples and vitro experiments confirm that EM alleviates NETs-induced glucocorticoid resistance via inhibiting PI3K/AKT, thereby repressing inflammation.
Conclusions: Our study provides a comprehensive proteogenomic characterization of how EM alleviates NET-related inflammation, and identify PI3K/AKT play a critical role in the mechanism by which EM inhibits inflammation.
Keywords: Erythromycin; Inflammation; NETs; PI3K/AKT; Proteogenomic.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The project titled “Neutrophil extracellular traps (NETs) enhances the glucocorticoid resistance of mononuclear cells exposed to CSE by upregulating activity of PI3K-δ/ Akt?”has been approved by the First Affiliated Hospital of Guangxi Medical University in 2018, and in accordance with the guidelines of the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (number: 2016-KY-048). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

