GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development
- PMID: 22883108
- PMCID: PMC3489871
- DOI: 10.1186/1471-2229-12-144
GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development
Abstract
Background: As a large family of regulatory proteins, WRKY transcription factors play essential roles in the processes of adaptation to diverse environmental stresses and plant growth and development. Although several studies have investigated the role of WRKY transcription factors during these processes, the mechanisms underlying the function of WRKY members need to be further explored, and research focusing on the WRKY family in cotton crops is extremely limited.
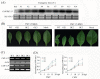
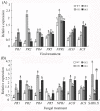
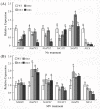
Results: In the present study, a gene encoding a putative WRKY family member, GhWRKY15, was isolated from cotton. GhWRKY15 is present as a single copy gene, and a transient expression analysis indicated that GhWRKY15 was localised to the nucleus. Additionally, a group of cis-acting elements associated with the response to environmental stress and plant growth and development were detected in the promoter. Consistently, northern blot analysis showed that GhWRKY15 expression was significantly induced in cotton seedlings following fungal infection or treatment with salicylic acid, methyl jasmonate or methyl viologen. Furthermore, GhWRKY15-overexpressing tobacco exhibited more resistance to viral and fungal infections compared with wild-type tobacco. The GhWRKY15-overexpressing tobacco also exhibited increased RNA expression of several pathogen-related genes, NONEXPRESSOR OF PR1, and two genes that encode enzymes involved in ET biosynthesis. Importantly, increased activity of the antioxidant enzymes POD and APX during infection and enhanced expression of NtAPX1 and NtGPX in transgenic tobacco following methyl viologen treatment were observed. Moreover, GhWRKY15 transcription was greater in the roots and stems compared with the expression in the cotyledon of cotton, and the stems of transgenic plants displayed faster elongation at the earlier shooting stages compared with wide type tobacco. Additionally, exposure to abiotic stresses, including cold, wounding and drought, resulted in the accumulation of GhWRKY15 transcripts.
Conclusion: Overall, our data suggest that overexpression of GhWRKY15 may contribute to the alteration of defence resistance to both viral and fungal infections, probably through regulating the ROS system via multiple signalling pathways in tobacco. It is intriguing that GhWRKY15 overexpression in tobacco affects plant growth and development, especially stem elongation. This finding suggests that the role of the WRKY proteins in disease resistance may be closely related to their function in regulating plant growth and development.
Figures





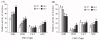


References
-
- Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa) J Integr Plant Biol. 2007;49:827–842. doi: 10.1111/j.1744-7909.2007.00504.x. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

