Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes
- PMID: 16905658
- PMCID: PMC1560908
- DOI: 10.1105/tpc.105.038844
Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes
Abstract
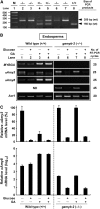
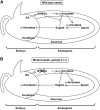
Expression of alpha-amylase genes during cereal grain germination and seedling growth is regulated negatively by sugar in embryos and positively by gibberellin (GA) in endosperm through the sugar response complex (SRC) and the GA response complex (GARC), respectively. We analyzed two alpha-amylase promoters, alphaAmy3 containing only SRC and alphaAmy8 containing overlapped SRC and GARC. alphaAmy3 was sugar-sensitive but GA-nonresponsive in both rice (Oryza sativa) embryos and endosperms, whereas alphaAmy8 was sugar-sensitive in embryos and GA-responsive in endosperms. Mutation of the GA response element (GARE) in the alphaAmy8 promoter impaired its GA response but enhanced sugar sensitivity, and insertion of GARE in the alphaAmy3 promoter rendered it GA-responsive but sugar-insensitive in endosperms. Expression of the GARE-interacting transcription factor MYBGA was induced by GA in endosperms, correlating with the endosperm-specific alphaAmy8 GA response. alphaAmy8 became sugar-sensitive in MYBGA knockout mutant endosperms, suggesting that the MYBGA-GARE interaction overrides the sugar sensitivity of alphaAmy8. In embryos overexpressing MYBGA, alphaAmy8 became sugar-insensitive, indicating that MYBGA affects sugar repression. alpha-Amylase promoters active in endosperms contain GARE, whereas those active in embryos may or may not contain GARE, confirming that the GARE and GA-induced MYBGA interaction prevents sugar feedback repression of endosperm alpha-amylase genes. We demonstrate that the MYBGA-GARE interaction affects sugar feedback control in balanced energy production during seedling growth and provide insight into the control mechanisms of tissue-specific regulation of alpha-amylase expression by sugar and GA signaling interference.
Figures






References
-
- Akazawa, T., and Hara-Mishimura, I. (1985). Topographic aspects of biosynthesis, extracellular secretion and intracellular storage of proteins in plant cells. Annu. Rev. Plant Physiol. 70 441–472.
-
- Beck, E., and Ziegler, P. (1989). Biosynthesis and degradation of starch in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40 95–117.
-
- Cercos, M., Gomez-Cadenas, A., and Ho, T.H. (1999). Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: Cis- and trans-acting elements involved in the co- ordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 19 107–118. - PubMed
-
- Chan, M.T., Chao, Y.C., and Yu, S.M. (1994). Novel gene expression system for plant cells based on induction of alpha-amylase promoter by carbohydrate starvation. J. Biol. Chem. 269 17635–17641. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

