Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA
- PMID: 33506766
- PMCID: PMC8274402
- DOI: 10.1016/j.annonc.2020.02.011
Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA
Abstract
Background: Early cancer detection could identify tumors at a time when outcomes are superior and treatment is less morbid. This prospective case-control sub-study (from NCT02889978 and NCT03085888) assessed the performance of targeted methylation analysis of circulating cell-free DNA (cfDNA) to detect and localize multiple cancer types across all stages at high specificity.
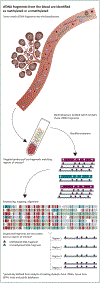
Participants and methods: The 6689 participants [2482 cancer (>50 cancer types), 4207 non-cancer] were divided into training and validation sets. Plasma cfDNA underwent bisulfite sequencing targeting a panel of >100 000 informative methylation regions. A classifier was developed and validated for cancer detection and tissue of origin (TOO) localization.
Results: Performance was consistent in training and validation sets. In validation, specificity was 99.3% [95% confidence interval (CI): 98.3% to 99.8%; 0.7% false-positive rate (FPR)]. Stage I-III sensitivity was 67.3% (CI: 60.7% to 73.3%) in a pre-specified set of 12 cancer types (anus, bladder, colon/rectum, esophagus, head and neck, liver/bile-duct, lung, lymphoma, ovary, pancreas, plasma cell neoplasm, stomach), which account for ∼63% of US cancer deaths annually, and was 43.9% (CI: 39.4% to 48.5%) in all cancer types. Detection increased with increasing stage: in the pre-specified cancer types sensitivity was 39% (CI: 27% to 52%) in stage I, 69% (CI: 56% to 80%) in stage II, 83% (CI: 75% to 90%) in stage III, and 92% (CI: 86% to 96%) in stage IV. In all cancer types sensitivity was 18% (CI: 13% to 25%) in stage I, 43% (CI: 35% to 51%) in stage II, 81% (CI: 73% to 87%) in stage III, and 93% (CI: 87% to 96%) in stage IV. TOO was predicted in 96% of samples with cancer-like signal; of those, the TOO localization was accurate in 93%.
Conclusions: cfDNA sequencing leveraging informative methylation patterns detected more than 50 cancer types across stages. Considering the potential value of early detection in deadly malignancies, further evaluation of this test is justified in prospective population-level studies.
Keywords: cancer; cell-free DNA; methylation; next-generation sequencing.
Copyright © 2020 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Disclosures The Mayo Clinic was compensated for MCL's advisory board activities for GRAIL, Inc. GRO reports personal fees from GRAIL, Inc. during the conduct of the study as well as personal fees from Inivata, Sysmex, AstraZeneca, Janssen, Illumina, and Foundation Medicine outside the submitted work. EAK reports personal fees from GRAIL, Inc. during the conduct of the study. MVS reports personal fees and other from McKesson and personal fees from GRAIL, Inc. during the conduct of the study as well as other from Merck and Bristol-Myers Squibb outside the submitted work. CS reports grants from Pfizer, AstraZeneca, BMS, Roche-Ventana, and Boehringer-Ingelheim; has consulted for Pfizer, Novartis, GlaxoSmithKline, MSD, BMS, Celgene, AstraZeneca, Illumina, Genentech, Roche-Ventana, GRAIL, Inc., Medicxi, and the Sarah Cannon Research Institute; has stock options of Apogen Biotechnologies, Epic Bioscience, GRAIL, Inc., and has stock options in and is co-founder of Achilles Therapeutics. CS is Royal Society Napier Research Professor. HA reports personal fees from GRAIL, Inc., during the conduct of the study as well as other from Illumina Inc. outside the submitted work; in addition, HA has patents pending to GRAIL, Inc. DAB reports grants from the National Cancer Institute, other from Berry Consultants, LLC, outside the submitted work. TCC reports personal fees and other from Illumina, Inc., outside the submitted work. CC reports personal fees and other (stock options) from GRAIL, Inc., during the conduct of the study as well as personal fees from Genentech outside the submitted work; in addition, CC has a patent pending outside the submitted work. KD reports personal fees from GRAIL, Inc. during the conduct of the study and other from Alphabet outside the submitted work. FJC reports research support from GRAIL, Inc. MD reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, MD has patents pending to GRAIL, Inc. SF reports personal fees and other from GRAIL, Inc., during the conduct of the study as well as personal fees and other from 23andMe and other from Illumina outside the submitted work. APF reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, APF has patents pending to GRAIL, Inc. DF reports personal fees from GRAIL, Inc. during the conduct of the study as well as personal fees from Roche Sequencing Solutions outside the submitted work; in addition, DF has patents pending to GRAIL, Inc. SG reports personal fees and other from GRAIL, Inc. during the conduct of the study as well as other from Illumina outside the submitted work. S. Gross reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, S. Gross has patents pending to GRAIL, Inc. MPH reports personal fees and other from GRAIL, Inc. during the conduct of the study as well as other from Jazz Pharmaceuticals and Natera outside the submitted work. SAP reports personal fees and other from GRAIL, Inc. during the conduct of the study as well as other from Natera, Inc. outside of the submitted work. EH reports personal fees and other from GRAIL, Inc. during the conduct of the study; in addition, EH has patents pending to GRAIL, Inc. NH reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, NH has patents pending to GRAIL, Inc. CH reports personal fees and other from GRAIL, Inc. during the conduct of the study as well as other from Illumina outside the submitted work. QL reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, QL has patents pending to GRAIL, Inc. AJ reports personal fees from GRAIL, Inc. during the conduct of the study and personal fees from Illumina outside the submitted work; in addition, AJ has patents pending to GRAIL, Inc. and a patent (differential tagging of RNA for preparation of a cell-free DNA/RNA sequencing library) issued to GRAIL, Inc. RK reports personal fees from GRAIL, Inc. during the conduct of the study as well as personal fees from Mindstrong Health, personal fees from Lyell Immunopharma, personal fees from LifeMine, personal fees from Wisdo, personal fees from Medical Creations/Extremity, and personal fees from FOG Pharma all outside the submitted work. KNK reports personal fees and other from GRAIL, Inc. during the conduct of the study as well as other from Illumina outside the submitted work. ML reports personal fees and other from GRAIL, Inc. during the conduct of the study as well as personal fees and other from Genentech, Inc., personal fees and other from Google Life Sciences, personal fees and other from Boreal Genomics, and personal fees and other from Genomic Health, Inc. outside the submitted work; in addition, ML has a patent arising from the CCGA work pending to GRAIL, Inc. MCM reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, MCM has patents pending to GRAIL, Inc. CM reports personal fees and other from GRAIL, Inc. during the conduct of the study; in addition, CM has a patent pending to GRAIL, Inc. VD reports personal fees and other from GRAIL, Inc. during the conduct of the study. JN reports personal fees from GRAIL Inc. during the conduct of the study as well as personal fees from Verily Life Sciences (formerly part of Google) outside the submitted work. Joshua N. reports personal fees and other from GRAIL, Inc. during the conduct of the study. VN reports personal fees from GRAIL Inc. during the conduct of the study; in addition, VN has patents pending to GRAIL, Inc. RVS reports personal fees from GRAIL, Inc. during the conduct of the study as well as personal fees from Guardant Health outside the submitted work; in addition, RVS has a patent pending to GRAIL, Inc. AS reports personal fees from GRAIL, Inc. during the conduct of the study and is the owner of Illumina stock. LS reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, LS has a patent pending to GRAIL, Inc. MS reports personal fees from Celgene, personal fees from Millenium/Takeda, and personal fees from Syros outside the submitted work. DS reports other from US Oncology during the conduct of the study. AV reports personal fees and other from GRAIL, Inc. during the conduct of the study as well as other from Illumina outside the submitted work. OV reports personal fees from GRAIL, Inc. during the conduct of the study; in addition, OV has patents pending to GRAIL, Inc. SRC reports research support from GRAIL, Inc. JY reports personal fees and other from GRAIL Inc. during the conduct of the study as well as personal fees and other from Acerta Pharma B.V., personal fees from Forty Seven Inc., other from BeiGene, Ltd., other from Celgene Corporation, other from Loxo Oncology, Inc., other from Nektar Therapeutics, other from Corvus Pharmaceuticals, Inc., and other from Illumina, Inc., all outside the submitted work. AB reports a financial interest in GRAIL, Inc. via Foresite Capital’s funds and personal equity. AMA is a founder, employee, and shareholder at GRAIL, Inc. and a paid advisor to Foresite Capital and Myst Therapeutics. JB reports personal fees from GRAIL, Inc. as well as patents pending to GRAIL, Inc. during the conduct of the study; JB also has patents issued to Roche and Philips Medical Systems outside of this work. PF reports personal fees from GRAIL, Inc. as well as patents pending to GRAIL, Inc. during the conduct of the study. WL reports personal fees and other from GRAIL, Inc. during the conduct of the study and personal fees from Genentech outside of this work. TM reports personal fees and other from GRAIL, Inc. and a patent pending to GRAIL, Inc. during the conduct of the study; TM also reports personal fees and other from Lexent Bio, HTG Molecular, and NDA Partners, as well as other from Genomic Health and personal fees from Terumo Medical outside of this work. RS, RL, TW, AS, ON, LZ, RC, CY, PS, NR, CC, AY, A. Shanmugam, JS, GA, AM, JZ, HC, GC, KCS, XC, BA, JL, JY, FA, LB, J. Berman, JC, TK, SB, JFB, CB, TCC, DC, ZD, ETF, A-RH, RJ, BJ, QL, SN, CN, SP, RR, OS, ES, AJS, SS, KKS, ST, JMT, RTW, XY, JY, and NZ report personal fees from GRAIL, Inc. during the conduct of the study. The remaining authors have declared no conflict of interest.
Figures




Comment in
-
Pan-Cancer Early Detection: Hype or Hope?Cancer Cell. 2020 Jul 13;38(1):23-24. doi: 10.1016/j.ccell.2020.05.021. Epub 2020 Jun 11. Cancer Cell. 2020. PMID: 32531269
References
-
- NIH National Cancer Insitute. Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 18 Regs Research Data, Nov 2017 Sub (1973-2015) National Cancer Institute, DCCPS, Surveillance Research Program, released April 2018, based on the November 2017 submission. Statistic based on all invasive cancers, ages 50+ at diagnosis. [www.seer.cancer.gov].
-
- Noone A, Howlander N, Krapcho M, et al., eds. SEER Cancer Statistics Review, 1975-2015, National Cancer Institute, software version 8.3.6. Bethesda. Available at: https://seer.cancer.gov/csr/1975_2015/.
-
- Narayan A, Fischer A, Zhang Z, et al. Nationwide cross-sectional adherence to mammography screening guidelines: national behavioral risk factor surveillance system survey results. Breast Cancer Res Treat. 2017;164(3):719–725. - PubMed
-
- Brasher P, Tanner N, Yeager D, Silvestri G. Adherence to annual lung cancer screening within the Veterans Health Administration lung cancer screening demonstration project. Chest. 2018;154(4):636A–637A. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

