Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study
- PMID: 27452608
- PMCID: PMC5014570
- DOI: 10.1016/S0140-6736(16)30826-1
Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study
Abstract
Background: Chronic injury in kidney transplants remains a major cause of allograft loss. The aim of this study was to identify a gene set capable of predicting renal allografts at risk of progressive injury due to fibrosis.
Methods: This Genomics of Chronic Allograft Rejection (GoCAR) study is a prospective, multicentre study. We prospectively collected biopsies from renal allograft recipients (n=204) with stable renal function 3 months after transplantation. We used microarray analysis to investigate gene expression in 159 of these tissue samples. We aimed to identify genes that correlated with the Chronic Allograft Damage Index (CADI) score at 12 months, but not fibrosis at the time of the biopsy. We applied a penalised regression model in combination with permutation-based approach to derive an optimal gene set to predict allograft fibrosis. The GoCAR study is registered with ClinicalTrials.gov, number NCT00611702.
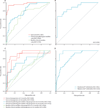
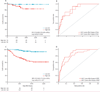
Findings: We identified a set of 13 genes that was independently predictive for the development of fibrosis at 1 year (ie, CADI-12 ≥2). The gene set had high predictive capacity (area under the curve [AUC] 0·967), which was superior to that of baseline clinical variables (AUC 0·706) and clinical and pathological variables (AUC 0·806). Furthermore routine pathological variables were unable to identify which histologically normal allografts would progress to fibrosis (AUC 0·754), whereas the predictive gene set accurately discriminated between transplants at high and low risk of progression (AUC 0·916). The 13 genes also accurately predicted early allograft loss (AUC 0·842 at 2 years and 0·844 at 3 years). We validated the predictive value of this gene set in an independent cohort from the GoCAR study (n=45, AUC 0·866) and two independent, publically available expression datasets (n=282, AUC 0·831 and n=24, AUC 0·972).
Interpretation: Our results suggest that this set of 13 genes could be used to identify kidney transplant recipients at risk of allograft loss before the development of irreversible damage, thus allowing therapy to be modified to prevent progression to fibrosis.
Funding: National Institutes of Health.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

Comment in
-
Transplant survival: knowing the future.Lancet. 2016 Sep 3;388(10048):940-1. doi: 10.1016/S0140-6736(16)30967-9. Epub 2016 Jul 22. Lancet. 2016. PMID: 27452609 No abstract available.
-
Transplantation: GoCAR - on the road to reduced allograft loss.Nat Rev Urol. 2016 Sep;13(9):495. doi: 10.1038/nrurol.2016.149. Epub 2016 Aug 9. Nat Rev Urol. 2016. PMID: 27502547 No abstract available.
-
Foretelling Graft Outcome by Molecular Evaluation of Renal Allograft Biopsies: The GoCAR Study.Transplantation. 2017 Jan;101(1):5-7. doi: 10.1097/TP.0000000000001512. Transplantation. 2017. PMID: 28005695 No abstract available.
-
Biopsy transcriptome expression profiling: proper validation is key.Lancet. 2017 Feb 11;389(10069):600-601. doi: 10.1016/S0140-6736(17)30282-9. Lancet. 2017. PMID: 28195055 No abstract available.
-
Knowing the allograft's destiny.Transl Androl Urol. 2017 Apr;6(2):313-314. doi: 10.21037/tau.2017.03.27. Transl Androl Urol. 2017. PMID: 28540243 Free PMC article. No abstract available.
References
-
- Matas AJ, Smith JM, Skeans MA, et al. OPTN/SRTR 2013 annual data report: kidney. Am J Transplant. 2015;15(suppl 2):1–34. - PubMed
-
- Meier-Kriesche HU, Schold JD, Srinivas TR, Kaplan B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4:378–383. - PubMed
-
- Wolfe RA, Roys EC, Merion RM. Trends in organ donation and transplantation in the United States, 1999–2008. Am J Transplant. 2010;10:961–972. - PubMed
-
- Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349:2326–2333. - PubMed
-
- Yilmaz S, McLaughlin K, Paavonen T, et al. Clinical predictors of renal allograft histopathology: a comparative study of single-lesion histology versus a composite, quantitative scoring system. Transplantation. 2007;83:671–676. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

