Randomized Phase II Trial of Dendritic Cell/Myeloma Fusion Vaccine with Lenalidomide Maintenance after Upfront Autologous Hematopoietic Cell Transplantation for Multiple Myeloma: BMT CTN 1401
- PMID: 37463058
- PMCID: PMC10690096
- DOI: 10.1158/1078-0432.CCR-23-0235
Randomized Phase II Trial of Dendritic Cell/Myeloma Fusion Vaccine with Lenalidomide Maintenance after Upfront Autologous Hematopoietic Cell Transplantation for Multiple Myeloma: BMT CTN 1401
Erratum in
-
Correction: Randomized Phase II Trial of Dendritic Cell/Myeloma Fusion Vaccine with Lenalidomide Maintenance after Upfront Autologous Hematopoietic Cell Transplantation for Multiple Myeloma: BMT CTN 1401.Clin Cancer Res. 2024 Oct 1;30(19):4542. doi: 10.1158/1078-0432.CCR-24-2408. Clin Cancer Res. 2024. PMID: 39350638 Free PMC article. No abstract available.
Abstract
Purpose: Vaccination with dendritic cell (DC)/multiple myeloma (MM) fusions has been shown to induce the expansion of circulating multiple myeloma-reactive lymphocytes and consolidation of clinical response following autologous hematopoietic cell transplant (auto-HCT).
Patients and methods: In this randomized phase II trial (NCT02728102), we assessed the effect of DC/MM fusion vaccination, GM-CSF, and lenalidomide maintenance as compared with control arms of GM-CSF and lenalidomide or lenalidomide maintenance alone on clinical response rates and induction of multiple myeloma-specific immunity at 1-year posttransplant.
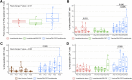
Results: The study enrolled 203 patients, with 140 randomized posttransplantation. Vaccine production was successful in 63 of 68 patients. At 1 year, rates of CR were 52.9% (vaccine) and 50% (control; P = 0.37, 80% CI 44.5%, 61.3%, and 41.6%, 58.4%, respectively), and rates of VGPR or better were 85.3% (vaccine) and 77.8% (control; P = 0.2). Conversion to CR at 1 year was 34.8% (vaccine) and 27.3% (control; P = 0.4). Vaccination induced a statistically significant expansion of multiple myeloma-reactive T cells at 1 year compared with before vaccination (P = 0.024) and in contrast to the nonvaccine arm (P = 0.026). Single-cell transcriptomics revealed clonotypic expansion of activated CD8 cells and shared dominant clonotypes between patients at 1-year posttransplant.
Conclusions: DC/MM fusion vaccination with lenalidomide did not result in a statistically significant increase in CR rates at 1 year posttransplant but was associated with a significant increase in circulating multiple myeloma-reactive lymphocytes indicative of tumor-specific immunity. Site-specific production of a personalized cell therapy with centralized product characterization was effectively accomplished in the context of a multicenter cooperative group study. See related commentary by Qazilbash and Kwak, p. 4703.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures



Comment in
-
Personalized Medicine's Coming of Age: One Drug, One Patient.Clin Cancer Res. 2023 Dec 1;29(23):4703-4705. doi: 10.1158/1078-0432.CCR-23-2194. Clin Cancer Res. 2023. PMID: 37733765
References
-
- Nakamura K, Smyth MJ, Martinet L. Cancer immunoediting and immune dysregulation in multiple myeloma. Blood 2020;136:2731–40. - PubMed
-
- Alrasheed N, Lee L, Ghorani E, Henry JY, Conde L, Chin M, et al. . Marrow-infiltrating regulatory T cells correlate with the presence of dysfunctional CD4+PD-1+ cells and inferior survival in patients with newly diagnosed multiple myeloma. Clin Cancer Res 2020;26:3443–54. - PubMed

