Autologous T cell therapy for PRAME+ advanced solid tumors in HLA-A*02+ patients: a phase 1 trial
- PMID: 40205198
- PMCID: PMC12283372
- DOI: 10.1038/s41591-025-03650-6
Autologous T cell therapy for PRAME+ advanced solid tumors in HLA-A*02+ patients: a phase 1 trial
Erratum in
-
Author Correction: Autologous T cell therapy for PRAME+ advanced solid tumors in HLA-A*02+ patients: a phase 1 trial.Nat Med. 2025 Jul;31(7):2453. doi: 10.1038/s41591-025-03731-6. Nat Med. 2025. PMID: 40301562 Free PMC article. No abstract available.
Abstract
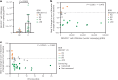
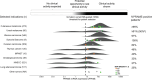
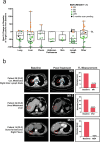
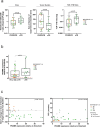
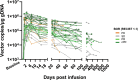
In contrast to chimeric antigen receptor T cells, T cell receptor (TCR)-engineered T cells can target intracellular tumor-associated antigens crucial for treating solid tumors. However, most trials published so far show limited clinical activity. Here we report interim data from a first-in-human, multicenter, open-label, 3 + 3 dose-escalation/de-escalation phase 1 trial studying IMA203, an autologous preferentially expressed antigen in melanoma (PRAME)-directed TCR T cell therapy in HLA-A*02+ patients with PRAME+ recurrent and/or refractory solid tumors, including melanoma and sarcoma. Primary objectives include the evaluation of safety and tolerability and the determination of the maximum tolerated dose (MTD) and/or recommended dose for extension. Secondary objectives include the evaluation of IMA203 TCR-engineered T cell persistence in peripheral blood, tumor response as well as duration of response. A total of 27 patients were enrolled in the phase 1a dose escalation and 13 patients in the phase 1b dose extension. IMA203 T cells were safe, and the MTD was not reached. Of the 41 patients receiving treatment (that is, who started lymphodepletion), severe cytokine release syndrome was observed in 4.9% (2/41), and severe neurotoxicity did not occur. In the 40 patients treated with IMA203, an overall response rate consisting of patients with unconfirmed or confirmed response (u/cORR) of 52.5% (21/40) and a cORR of 28.9% (11/38) was observed with a median duration of response of 4.4 months (range, 2.4-23.0, 95% confidence interval: 2.6-not reached) across multiple indications. Rapid T cell engraftment and long-term persistence of IMA203 T cells were observed. IMA203 T cells trafficked to all organs, and confirmed responses were more frequent in patients with higher dose. T cell exhaustion was not observed in the periphery; deep responses were enriched at higher PRAME expression; and higher T cell infiltration resulted in longer progression-free survival. Overall, IMA203 showed promising anti-tumor activity in multiple solid tumors, including refractory melanoma. ClinicalTrials.gov identifier: NCT03686124 .
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.W.: honoraria: Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Eli Lilly, GWT, Janssen, Merck Novartis, Pfizer, Serono and Synlab; consulting or advisory role: Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo Europe GmbH, Eli Lilly, ImCheck Therapeutics, Immatics, ISA Pharmaceuticals, Novartis, PharmaMar, Regeneron, Tacalyx and Zymeworks; research funding (to institution): Roche; travel, accommodations, expenses: Amgen, AstraZeneca, Bristol Myers Squibb, Daiichi Sankyo Europe GmbH, GEMoaB, Immatics, Iovance Biotherapeutics, Janssen Oncology, Merck/Senoro, Pfizer and Sanofi/Aventis. D.M.A.: research funding: Adaptimmune, GlaxoSmithKline and Immatics. A.M.T.: consulting or advisory role: Avstera, BioEclipse Therapeutics, BrYet, Diaccurate, Macrogenics, Nex-I and Vincerx; research funding: BrYet (to institution), AbbVie, Agenus, Immatics, Macrogenics, Novocure Ltd., OBI Pharma, Orionis, Parker Institute for Cancer Immunotherapy, Tachyon, Tempus, Tvardi Therapeutics and Vividion Therapeutics. T.A.W.H.: honoraria: Amgen, Bristol Myers Squibb, GlaxoSmithKline and Jazz Pharmaceuticals; consulting or advisory role: AbbVie, Amgen, Bristol Myers Squibb, GlaxoSmithKline, Janssen, Jazz Pharmaceuticals, Kite/Gilead, Novartis, Otsuka, Pfizer, Pierre-Fabre and Sanofi; travel, accomodations, expenses: AbbVie, Amgen, Astellas, BeiGene, Bristol Myers Squibb, GlaxoSmithKline, Immatics, Janssen, Jazz Pharmaceuticals, Kite/Gilead, Kyverna Therapeutics, Medac, Neovii, Sanofi and Sobi. A.A.J.: stock or other ownership: Avenge Bio; consulting or advisory role: Avenge Bio, Eisei, GLG, Guidepoint, Macrogenics, Sentinel Bio and Theolytics; research funding: AstraZeneca, Eli Lilly, Immatics, Immunon, Iovance, Macrogenics, Merck and Xencor; travel, accomodations, expenses: Striker. R.R.: consulting or advisory role: Allogene, Autolus, Gilead, Incyte, Orca Bio, Quell Biotherapeutics, Sana Biotechnology and TScan; research funding (all to institution): AbbVie, Atara Biotherapeutics, Bristol Myers Squibb, Cabaletta, CareDx, Genentech, Gilead, Immatics, Incyte, Johnson & Johnson, Precision Biosciences, Sanofi, Synthekine, Takeda, TCR2 and TScan; expert testimony: Bayer; travel, accommodations, expenses: Gilead and Johnson & Johnson. C.B.: consulting fees: AOK Germany, AstraZeneca, Bayer Healthcare, BioNTech, Lindis Biotech and Sanofi Aventis; payment or honoraria for lectures, presentations, speakersʼ bureaus, manuscript writing or educational events: med update, Merck Serono and Roche Pharma; data safety monitoring board or advisory board: German Society of Hematology and Oncology (DGHO), Hamburg Cancer Society (HKG), German Cancer Society (DKG), National Network of German Cancer Centers (CCC)/German Cancer Aid (DKH) and Northern Germany Society of Internal Medicine (NWGIM). W.A.: consulting or advisory role: Janssen; research funding (received through institution): Affimed and BioNTech; travel, accommodation, expenses: BioNTech, Immatics and Janssen; honoraria: Astellas, AstraZeneca, GlaxoSmithKline and Janssen. H.-G.R.: stock or other ownership: CureVac, Immatics and ViferaXS; consulting or advisory role: Immatics; patents, royalities, other intellectual property: CureVac, Immatics and ViferaXS. P.B.: consulting or advisory role: BeiGene, Bristol Myers Squibb and Kite; research funding: Bristol Myers Squibb. J.J.L.: data safety and monitoring board: AbbVie, Agenus, Evaxion, Immutep and Shionogi; scientific advisory board (no stock): 7 Hills, Affivant, BioCytics, Bright Peak, Exo, Fstar, Inzen, RefleXion and Xilio; scientific advisory board (stock): Actym, Alphamab Oncology, Arch Oncology, Duke Street Bio, Elipscience, Kanaph, NeoTx, Onc.AI, OncoNano, physIQ, Pyxis, Saros, Stipe and Tempest; consultancy with compensation: AbbVie, Agenus, Alnylam, AstraZeneca, Askgene, Atomwise, Bayer, Bristol Myers Squibb, Castle, Checkmate, Codiak, Crown, Cugene, Curadev, Day One, Eisai, EMD Serono, Endeavor, Flame, G1 Therapeutics, Genentech, Geneos, Gilead, Glenmark, HotSpot, Kadmon, Ko Bio Labs, Krystal, KSQ, Janssen, Ikena, Inzen, Immatics, Immunocore, Incyte, Instil, IO Biotech, LegoChem, Lyvgen, Macrogenics, Merck, Mersana, Nektar, Novartis, Partner, Pfizer, Pioneering Medicines, PsiOxus, Regeneron, Replimmune, Ribon, Roivant, Servier, STINGthera, Storm, Sumoitomo, Synlogic, Synthekine and Teva; research support (all to institution): AbbVie, Astellas, AstraZeneca, Bristol Myers Squibb, Corvus, Day One, EMD Serono, Fstar, Genmab, Hot Spot, Ikena, Immatics, Imugene, Incyte, Janux, Kadmon, KAHR, Macrogenics, Merck, Synlogic, Takeda, Trishula, Tizona, Tscan, Werewolf and Xencor; patents: US-11638728 (‘Microbiome biomarkers for anti-PD-1/PD-L1 responsiveness: diagnostic, prognostic and therapeutic uses thereof’). K.A., L.B., S.B., J.F., S.G., S.H., N.H., J.H., M.A.K., D.M., A.M.-M., R.M., M.L., H.S., O.S., C.W., C.R., H.S.-J., T.W. and C.M.B. were employees of Immatics Biotechnologies GmbH during the course of this work and may have securities from Immatics. M.B.H., M.K., D.K., A.M., K.P., A.S. and S.W. were employees of Immatics US, Inc. during the course of this work and may have securities from Immatics. The other authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

