Allogeneic, off-the-shelf, SARS-CoV-2-specific T cells (ALVR109) for the treatment of COVID-19 in high-risk patients
- PMID: 36373249
- PMCID: PMC10316279
- DOI: 10.3324/haematol.2022.281946
Allogeneic, off-the-shelf, SARS-CoV-2-specific T cells (ALVR109) for the treatment of COVID-19 in high-risk patients
Abstract
Defects in T-cell immunity to SARS-CoV-2 have been linked to an increased risk of severe COVID-19 (even after vaccination), persistent viral shedding and the emergence of more virulent viral variants. To address this T-cell deficit, we sought to prepare and cryopreserve banks of virus-specific T cells, which would be available as a partially HLA-matched, off-the-shelf product for immediate therapeutic use. By interrogating the peripheral blood of healthy convalescent donors, we identified immunodominant and protective T-cell target antigens, and generated and characterized polyclonal virus-specific T-cell lines with activity against multiple clinically important SARS-CoV-2 variants (including 'delta' and 'omicron'). The feasibility of making and safely utilizing such virus-specific T cells clinically was assessed by administering partially HLA-matched, third-party, cryopreserved SARS-CoV-2-specific T cells (ALVR109) in combination with other antiviral agents to four individuals who were hospitalized with COVID-19. This study establishes the feasibility of preparing and delivering off-the-shelf, SARS-CoV-2-directed, virus-specific T cells to patients with COVID-19 and supports the clinical use of these products outside of the profoundly immune compromised setting (ClinicalTrials.gov number, NCT04401410).
Figures
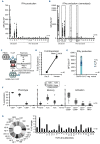
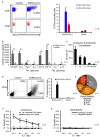


References
-
- Liao M, Liu Y, Yuan J, et al. . Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842-844. - PubMed
Publication types
MeSH terms
Supplementary concepts
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

