The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases
- PMID: 12917423
- PMCID: PMC8060950
- DOI: 10.1074/jbc.C300328200
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases
Abstract
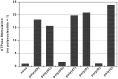
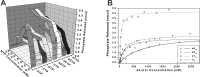
The putative NTPase/helicase protein from severe acute respiratory syndrome coronavirus (SARS-CoV) is postulated to play a number of crucial roles in the viral life cycle, making it an attractive target for anti-SARS therapy. We have cloned, expressed, and purified this protein as an N-terminal hexahistidine fusion in Escherichia coli and have characterized its helicase and NTPase activities. The enzyme unwinds double-stranded DNA, dependent on the presence of a 5' single-stranded overhang, indicating a 5'o 3' polarity of activity, a distinct characteristic of coronaviridae helicases. We provide the first quantitative analysis of the polynucleic acid binding and NTPase activities of a Nidovirus helicase, using a high throughput phosphate release assay that will be readily adaptable to the future testing of helicase inhibitors. All eight common NTPs and dNTPs were hydrolyzed by the SARS helicase in a magnesium-dependent reaction, stimulated by the presence of either single-stranded DNA or RNA. The enzyme exhibited a preference for ATP, dATP, and dCTP over the other NTP/dNTP substrates. Homopolynucleotides significantly stimulated the ATPase activity (15-25-fold) with the notable exception of poly(G) and poly(dG), which were non-stimulatory. We found a large variation in the apparent strength of binding of different homopolynucleotides, with dT24 binding over 10 times more strongly than dA24 as observed by the apparent Km.
Figures

References
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. - PubMed
-
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

