Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair
- PMID: 25533843
- PMCID: PMC4274264
- DOI: 10.1083/jcb.201405077
Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair
Abstract
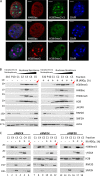
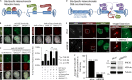
Heterochromatin is a barrier to DNA repair that correlates strongly with elevated somatic mutation in cancer. CHD class II nucleosome remodeling activity (specifically CHD3.1) retained by KAP-1 increases heterochromatin compaction and impedes DNA double-strand break (DSB) repair requiring Artemis. This obstruction is alleviated by chromatin relaxation via ATM-dependent KAP-1S824 phosphorylation (pKAP-1) and CHD3.1 dispersal from heterochromatic DSBs; however, how heterochromatin compaction is actually adjusted after CHD3.1 dispersal is unknown. In this paper, we demonstrate that Artemis-dependent DSB repair in heterochromatin requires ISWI (imitation switch)-class ACF1-SNF2H nucleosome remodeling. Compacted chromatin generated by CHD3.1 after DNA replication necessitates ACF1-SNF2H-mediated relaxation for DSB repair. ACF1-SNF2H requires RNF20 to bind heterochromatic DSBs, underlies RNF20-mediated chromatin relaxation, and functions downstream of pKAP-1-mediated CHD3.1 dispersal to enable DSB repair. CHD3.1 and ACF1-SNF2H display counteractive activities but similar histone affinities (via the plant homeodomains of CHD3.1 and ACF1), which we suggest necessitates a two-step dispersal and recruitment system regulating these opposing chromatin remodeling activities during DSB repair.
© 2014 Klement et al.
Figures








References
-
- Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A.A., Krempler A., Jeggo P.A., and Löbrich M.. 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28:3413–3427 10.1038/emboj.2009.276 - DOI - PMC - PubMed
-
- Brunton H., Goodarzi A.A., Noon A.T., Shrikhande A., Hansen R.S., Jeggo P.A., and Shibata A.. 2011. Analysis of human syndromes with disordered chromatin reveals the impact of heterochromatin on the efficacy of ATM-dependent G2/M checkpoint arrest. Mol. Cell. Biol. 31:4022–4035 10.1128/MCB.05289-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

