Distinct expression of interferon-induced protein with tetratricopeptide repeats (IFIT) 1/2/3 and other antiviral genes between subsets of dendritic cells induced by dengue virus 2 infection
- PMID: 27135915
- PMCID: PMC4948034
- DOI: 10.1111/imm.12615
Distinct expression of interferon-induced protein with tetratricopeptide repeats (IFIT) 1/2/3 and other antiviral genes between subsets of dendritic cells induced by dengue virus 2 infection
Abstract
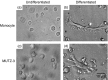
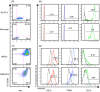
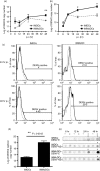
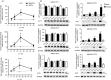
Dengue virus (DENV) infection is an emerging public health hazard threatening inhabitants of the tropics and sub-tropics. Dendritic cells (DCs) are one of the major targets of DENV and the initiators of the innate immune response against the virus. However, current in vitro research on the DENV-DC interaction is hampered by the low availability of ex vivo DCs and donor variation. In the current study, we attempted to develop a novel in vitro DC model using immature DCs derived from the myeloid leukaemia cell line MUTZ-3 (IMDCs) to investigate the DENV-DC interaction. The IMDCs morphologically and phenotypically resembled human immature monocyte-derived dendritic cells (IMMoDCs). However, the permissiveness of IMDCs to DENV2 was lower than that of IMMoDCs. RT-PCR arrays showed that a group of type I interferon (IFN) -inducible genes, especially IFIT1, IFITM1, and IFI27, were significantly up-regulated in IMMoDCs but not in IMDCs after DENV2 infection. Further investigation revealed that IFIT genes were spontaneously expressed at both transcriptional and protein levels in the naive IMDCs but not in the naive IMMoDCs. It is possible that the poor permissiveness of IMDCs to DENV2 was a result of the high basal levels of IFIT proteins. We conclude that the IMDC model, although less permissive to DENV2, is a useful platform for studying the suppression mechanism of DENV2 and we expand the knowledge of cellular factors that modulate DENV2 infection in the human body.
Keywords: MUTZ-3; dendritic cells; dengue virus.
© 2016 John Wiley & Sons Ltd.
Figures

References
-
- Halstead SB. Dengue. Lancet 2007; 370:1644–52. - PubMed
-
- World Health Organization . Dengue and severe dengue. Geneva: World Health Organization, 2012.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

