Interfering residues narrow the spectrum of MLV restriction by human TRIM5alpha
- PMID: 18166079
- PMCID: PMC2156100
- DOI: 10.1371/journal.ppat.0030200
Interfering residues narrow the spectrum of MLV restriction by human TRIM5alpha
Abstract
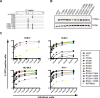
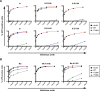
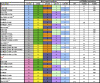
TRIM5alpha is a restriction factor that limits infection of human cells by so-called N- but not B- or NB-tropic strains of murine leukemia virus (MLV). Here, we performed a mutation-based functional analysis of TRIM5alpha-mediated MLV restriction. Our results reveal that changes at tyrosine(336) of human TRIM5alpha, within the variable region 1 of its C-terminal PRYSPRY domain, can expand its activity to B-MLV and to the NB-tropic Moloney MLV. Conversely, we demonstrate that the escape of MLV from restriction by wild-type or mutant forms of huTRIM5alpha can be achieved through interdependent changes at positions 82, 109, 110, and 117 of the viral capsid. Together, our results support a model in which TRIM5alpha-mediated retroviral restriction results from the direct binding of the antiviral PRYSPRY domain to the viral capsid, and can be prevented by interferences exerted by critical residues on either one of these two partners.
Conflict of interest statement
Figures





References
-
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. - PubMed
-
- Lilly F. Susceptibility to two strains of Friend leukemia virus in mice. Science. 1967;155:461–462. - PubMed
-
- Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
-
- Kozak CA, Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225:300–305. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

