Genomic analysis of mouse retinal development
- PMID: 15226823
- PMCID: PMC439783
- DOI: 10.1371/journal.pbio.0020247
Genomic analysis of mouse retinal development
Abstract
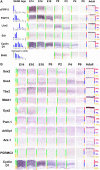
The vertebrate retina is comprised of seven major cell types that are generated in overlapping but well-defined intervals. To identify genes that might regulate retinal development, gene expression in the developing retina was profiled at multiple time points using serial analysis of gene expression (SAGE). The expression patterns of 1,051 genes that showed developmentally dynamic expression by SAGE were investigated using in situ hybridization. A molecular atlas of gene expression in the developing and mature retina was thereby constructed, along with a taxonomic classification of developmental gene expression patterns. Genes were identified that label both temporal and spatial subsets of mitotic progenitor cells. For each developing and mature major retinal cell type, genes selectively expressed in that cell type were identified. The gene expression profiles of retinal Müller glia and mitotic progenitor cells were found to be highly similar, suggesting that Müller glia might serve to produce multiple retinal cell types under the right conditions. In addition, multiple transcripts that were evolutionarily conserved that did not appear to encode open reading frames of more than 100 amino acids in length ("noncoding RNAs") were found to be dynamically and specifically expressed in developing and mature retinal cell types. Finally, many photoreceptor-enriched genes that mapped to chromosomal intervals containing retinal disease genes were identified. These data serve as a starting point for functional investigations of the roles of these genes in retinal development and physiology.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures





References
-
- Alexiades MR, Cepko CL. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. - PubMed
-
- Alexiades MR, Cepko CL. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development. 1997;124:1119–1131. - PubMed
-
- Altshuler D, Lo Turco JJ, Rush J, Cepko C. Taurine promotes the differentiation of a vertebrate retinal cell type in vitro. Development. 1993;119:1317–1328. - PubMed
-
- Anderson DJ. Lineages and transcription factors in the specification of vertebrate primary sensory neurons. Curr Opin Neurobiol. 1999;9:517–524. - PubMed
-
- Austin CP, Feldman DE, Ida JA, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

