Regulation and processing of maize histone deacetylase Hda1 by limited proteolysis
- PMID: 12897261
- PMCID: PMC167178
- DOI: 10.1105/tpc.013995
Regulation and processing of maize histone deacetylase Hda1 by limited proteolysis
Abstract
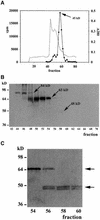
A maize histone deacetylase gene was identified as a homolog of yeast Hda1. The predicted protein corresponds to a previously purified maize deacetylase that is active as a protein monomer with a molecular weight of 48,000 and is expressed in all tissues of germinating embryos. Hda1 is synthesized as an enzymatically inactive protein with an apparent molecular weight of 84,000 that is processed to the active 48-kD form by proteolytic removal of the C-terminal part, presumably via a 65-kD intermediate. The enzymatically inactive 84-kD protein also is part of a 300-kD protein complex of unknown function. The proteolytic cleavage of ZmHda1 is regulated during maize embryo germination in vivo. Expression of the recombinant full-length protein and the 48-kD form confirmed that only the smaller enzyme form is active as a histone deacetylase. In line with this finding, we show that the 48-kD protein is able to repress transcription efficiently in a reporter gene assay, whereas the full-length protein, including the C-terminal part, lacks full repression activity. This report on the processing of Hda1-p84 to enzymatically active Hda1-p48 demonstrates that proteolytic cleavage is a mechanism to regulate the function of Rpd3/Hda1-type histone deacetylases.
Figures




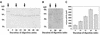


References
-
- Bertos, N.R., Wang, A.H., and Yang, Y.J. (2001). Class II histone deacetylases: Structure, functions, and regulations. Biochem. Cell Biol. 79, 243–252. - PubMed
-
- Bohner, S., Lenk, I.I., Rieping, M., Herold, M., and Gatz, C. (1999). Technical advances: Transcriptional activator TGV mediates dexamethasone-inducible and tetracycline-inactivable gene expression. Plant J. 19, 87–95. - PubMed
-
- Borchers, C., Peter, J.F., Hall, M.C., Kunkel, T.A., and Tomer, K.B. (2000). Identification of in-gel digested proteins by complementary peptide mass fingerprinting and tandem mass spectrometry data obtained on an electrospray ionization quadrupole time-of-flight mass spectrometer. Anal. Chem. 72, 1163–1168. - PubMed
-
- Boyer, L.A., Logie, C., Bonte, E., Becker, P.B., Wade, P.A., Wolffe, A.P., Wu, C., Imbalzano, A.N., and Peterson, C.L. (2000). Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 275, 18864–18870. - PubMed
-
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

