LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis
- PMID: 19448041
- PMCID: PMC2705037
- DOI: 10.1104/pp.109.136812
LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis
Erratum in
-
CORRECTION: Vol. 150: 1260-1271, 2009.Plant Physiol. 2017 Feb;173(2):1522-1523. doi: 10.1104/pp.16.01911. Plant Physiol. 2017. PMID: 28154116 Free PMC article. No abstract available.
Abstract
To gain insight into the molecular mechanism of RNA editing, we have characterized the low psii accumulation66 (lpa66) Arabidopsis (Arabidopsis thaliana) mutant, which displays a high chlorophyll fluorescence phenotype. Its perturbed chlorophyll fluorescence is reflected in reduced levels of photosystem II (PSII) proteins. In vivo protein labeling showed that synthesis rates of the PSII reaction center protein D1/D2 were lower, and turnover rates of PSII core proteins higher, than in wild-type counterparts. The assembly of newly synthesized proteins into PSII occurs in the lpa66 mutant but with reduced efficiency compared with the wild type. LPA66 encodes a chloroplast protein of the pentatricopeptide repeat family. In lpa66 mutants, editing of psbF that converts serine to phenylalanine is specifically impaired. Thus, LPA66 is specifically required for editing the psbF transcripts in Arabidopsis, and the amino acid alternation due to lack of editing strongly affects the efficiency of the assembly of PSII complexes.
Figures
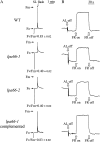
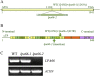




References
-
- Andrés C, Lurin C, Small ID (2007) The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol Plant 129 14–22
-
- Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572 - PubMed
-
- Bock R (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie 82 549–557 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

