Functional alignment of regulatory networks: a study of temperate phages
- PMID: 16477325
- PMCID: PMC1317652
- DOI: 10.1371/journal.pcbi.0010074
Functional alignment of regulatory networks: a study of temperate phages
Abstract
The relationship between the design and functionality of molecular networks is now a key issue in biology. Comparison of regulatory networks performing similar tasks can provide insights into how network architecture is constrained by the functions it directs. Here, we discuss methods of network comparison based on network architecture and signaling logic. Introducing local and global signaling scores for the difference between two networks, we quantify similarities between evolutionarily closely and distantly related bacteriophages. Despite the large evolutionary separation between phage lambda and 186, their networks are found to be similar when difference is measured in terms of global signaling. We finally discuss how network alignment can be used to pinpoint protein similarities viewed from the network perspective.
Conflict of interest statement
Figures

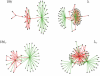
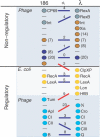
 . The numbers in the parentheses indicate multiple equivalent proteins, making the sum of all shown signaling differences equal to 2 × 43. The key regulators RecA, LexA, and CI are identified correctly, whereas the misidentification of CII with CIII is reasonable since both favor entry into lysogeny through the same pathway. The major discrepancy is associated with the different roles of Cro and Apl during lysis (the weak links from Cro to Q and N in λ).
. The numbers in the parentheses indicate multiple equivalent proteins, making the sum of all shown signaling differences equal to 2 × 43. The key regulators RecA, LexA, and CI are identified correctly, whereas the misidentification of CII with CIII is reasonable since both favor entry into lysogeny through the same pathway. The major discrepancy is associated with the different roles of Cro and Apl during lysis (the weak links from Cro to Q and N in λ).References
-
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. - PubMed
-
- Shen-Orr S, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. - PubMed
-
- Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

